Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
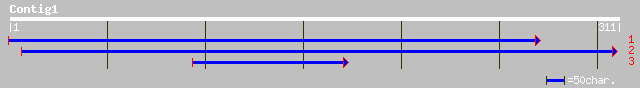
Query= KMC003407A_C01 KMC003407A_c01
(311 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_181799.2| hypothetical protein; protein id: At2g42710.1, ... 34 0.44
pir||S65148 tRNA adenylyltransferase (EC 2.7.7.25) - white lupin... 32 1.7
ref|NP_179007.1| CHP-rich zinc finger protein, putative; protein... 32 1.7
gb|EAA24575.1| hypothetical protein [Fusobacterium nucleatum sub... 32 2.8
gb|EAA22410.1| hypothetical protein [Plasmodium yoelii yoelii] 31 4.8
>ref|NP_181799.2| hypothetical protein; protein id: At2g42710.1, supported by cDNA:
gi_20268739, supported by cDNA: gi_21280898 [Arabidopsis
thaliana] gi|20268740|gb|AAM14073.1| unknown protein
[Arabidopsis thaliana] gi|21280899|gb|AAM45043.1|
unknown protein [Arabidopsis thaliana]
gi|26451659|dbj|BAC42926.1| unknown protein [Arabidopsis
thaliana]
Length = 415
Score = 34.3 bits (77), Expect = 0.44
Identities = 15/24 (62%), Positives = 20/24 (82%)
Frame = -2
Query: 310 MGPGFPVSIQSLSKAADNYKKMHV 239
MG G+PVSIQSLS+AAD + K+ +
Sbjct: 391 MGKGYPVSIQSLSRAADLHTKLQL 414
>pir||S65148 tRNA adenylyltransferase (EC 2.7.7.25) - white lupine
gi|1139585|gb|AAB03077.1| tRNA nucleotidyltransferase
Length = 560
Score = 32.3 bits (72), Expect = 1.7
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = -1
Query: 128 ICGNKKVKNLTYLCELDLFLIVF 60
I GN+ VK +TY+C+L +F IVF
Sbjct: 296 ISGNQPVKAMTYICDLTIFWIVF 318
>ref|NP_179007.1| CHP-rich zinc finger protein, putative; protein id: At2g13900.1
[Arabidopsis thaliana] gi|25411525|pir||G84511
hypothetical protein At2g13900 [imported] - Arabidopsis
thaliana gi|20197610|gb|AAM15153.1| hypothetical protein
[Arabidopsis thaliana]
Length = 661
Score = 32.3 bits (72), Expect = 1.7
Identities = 20/53 (37%), Positives = 27/53 (50%)
Frame = -1
Query: 209 VFQHESKFDKHVL*FFVHYH*FSLSCDICGNKKVKNLTYLCELDLFLIVFWCI 51
+F SK H+L FF SL CD+CG K TY+C +F++ CI
Sbjct: 232 IFIDHSKRHPHLLTFFPKQA--SLLCDVCGLVKESFPTYVCVRCIFVVHRDCI 282
>gb|EAA24575.1| hypothetical protein [Fusobacterium nucleatum subsp. vincentii ATCC
49256]
Length = 585
Score = 31.6 bits (70), Expect = 2.8
Identities = 19/46 (41%), Positives = 24/46 (51%)
Frame = +2
Query: 44 KVIYTKRRSETNQARTNMLNFSLSYCHIYHRTKKINGNEQKTTTHV 181
K+ YT R E NMLN LS +IY + KI+ NE T T +
Sbjct: 339 KIFYTMREEEVLSELKNMLN-QLSDRNIYSKLNKISKNEISTYTTI 383
>gb|EAA22410.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 1549
Score = 30.8 bits (68), Expect = 4.8
Identities = 18/68 (26%), Positives = 29/68 (42%)
Frame = +1
Query: 46 SDIHQKTIRNKSSSHKYVKFFTFLLPHISQDKENQW**TKNYNTCLSNFDSC*KTIASPY 225
S+ T+ + + H Y + P+++ D N NY N+D K + Y
Sbjct: 1335 SNRSNNTVNHMNDHHIYNPMYNLKFPYVNNDILNYNYNLANYQNY--NYDKTNKNMFKRY 1392
Query: 226 PRFIPHAF 249
P FIP+ F
Sbjct: 1393 PNFIPNNF 1400
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 255,879,647
Number of Sequences: 1393205
Number of extensions: 4909744
Number of successful extensions: 10761
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 10591
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10759
length of database: 448,689,247
effective HSP length: 79
effective length of database: 338,626,052
effective search space used: 8127025248
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)