Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003395A_C01 KMC003395A_c01
(539 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
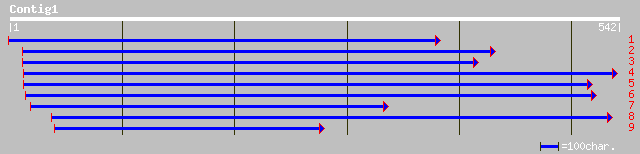
emb|CAC81075.1| dihydrofolate synthetase [Arabidopsis thaliana] 120 1e-26
ref|NP_187627.1| putative folylpolyglutamate synthetase; protein... 120 1e-26
emb|CAC82079.1| folylpolyglutamate-dihydrofolate synthetase [Ara... 73 2e-12
ref|NP_680131.1| tetrahydrofolylpolyglutamate synthase precursor... 73 2e-12
gb|AAM64329.1| tetrahydrofolylpolyglutamate synthase precursor-l... 73 2e-12
>emb|CAC81075.1| dihydrofolate synthetase [Arabidopsis thaliana]
Length = 625
Score = 120 bits (300), Expect = 1e-26
Identities = 63/114 (55%), Positives = 78/114 (68%), Gaps = 1/114 (0%)
Frame = -3
Query: 534 IPSDLGGIDLSWQFNLQRVWEKIMHGKAVTPSLENDFKISSKETLPPHEFLYDNVSN-GF 358
IPSD DL+WQF LQR+WEK + G L++ K LPPH+FL + G
Sbjct: 514 IPSDTRRKDLTWQFRLQRLWEKSIQG--TDAGLDHTLKPDGITALPPHDFLCGDAPQCGG 571
Query: 357 PSHNYSARSAVIPSLPLTIRWLRDCVKEHPSIRLQVLVTGSLHLVGDVLKLLKR 196
P+ SAV+PSLPLTI WLRDCV+ +PS++L+VLVTGSLHLVGDVL+LLKR
Sbjct: 572 PAGTPVTSSAVMPSLPLTINWLRDCVRRNPSLKLEVLVTGSLHLVGDVLRLLKR 625
>ref|NP_187627.1| putative folylpolyglutamate synthetase; protein id: At3g10160.1
[Arabidopsis thaliana]
gi|6143861|gb|AAF04408.1|AC010927_1 putative
folylpolyglutamate synthetase [Arabidopsis thaliana]
Length = 442
Score = 120 bits (300), Expect = 1e-26
Identities = 63/114 (55%), Positives = 78/114 (68%), Gaps = 1/114 (0%)
Frame = -3
Query: 534 IPSDLGGIDLSWQFNLQRVWEKIMHGKAVTPSLENDFKISSKETLPPHEFLYDNVSN-GF 358
IPSD DL+WQF LQR+WEK + G L++ K LPPH+FL + G
Sbjct: 331 IPSDTRRKDLTWQFRLQRLWEKSIQG--TDAGLDHTLKPDGITALPPHDFLCGDAPQCGG 388
Query: 357 PSHNYSARSAVIPSLPLTIRWLRDCVKEHPSIRLQVLVTGSLHLVGDVLKLLKR 196
P+ SAV+PSLPLTI WLRDCV+ +PS++L+VLVTGSLHLVGDVL+LLKR
Sbjct: 389 PAGTPVTSSAVMPSLPLTINWLRDCVRRNPSLKLEVLVTGSLHLVGDVLRLLKR 442
>emb|CAC82079.1| folylpolyglutamate-dihydrofolate synthetase [Arabidopsis thaliana]
Length = 492
Score = 73.2 bits (178), Expect = 2e-12
Identities = 40/104 (38%), Positives = 58/104 (55%)
Frame = -3
Query: 507 LSWQFNLQRVWEKIMHGKAVTPSLENDFKISSKETLPPHEFLYDNVSNGFPSHNYSARSA 328
+SWQF LQR+WE + G+A + S ++D K +E +S
Sbjct: 413 MSWQFGLQRIWESLARGEAKSNS-KSDSKGKEEE-----------------------KSF 448
Query: 327 VIPSLPLTIRWLRDCVKEHPSIRLQVLVTGSLHLVGDVLKLLKR 196
V SLP+ + WLRD ++ +R QVLVTGSLHLVGD+L+ +K+
Sbjct: 449 VFSSLPVAVDWLRDNARQSKQVRFQVLVTGSLHLVGDLLRFIKK 492
>ref|NP_680131.1| tetrahydrofolylpolyglutamate synthase precursor - like protein;
protein id: At3g55630.1, supported by cDNA: 230791.
[Arabidopsis thaliana]
Length = 470
Score = 73.2 bits (178), Expect = 2e-12
Identities = 40/104 (38%), Positives = 58/104 (55%)
Frame = -3
Query: 507 LSWQFNLQRVWEKIMHGKAVTPSLENDFKISSKETLPPHEFLYDNVSNGFPSHNYSARSA 328
+SWQF LQR+WE + G+A + S ++D K +E +S
Sbjct: 391 MSWQFGLQRIWESLARGEAKSNS-KSDSKGKEEE-----------------------KSF 426
Query: 327 VIPSLPLTIRWLRDCVKEHPSIRLQVLVTGSLHLVGDVLKLLKR 196
V SLP+ + WLRD ++ +R QVLVTGSLHLVGD+L+ +K+
Sbjct: 427 VFSSLPVAVDWLRDNARQSKQVRFQVLVTGSLHLVGDLLRFIKK 470
>gb|AAM64329.1| tetrahydrofolylpolyglutamate synthase precursor-like protein
[Arabidopsis thaliana]
Length = 470
Score = 73.2 bits (178), Expect = 2e-12
Identities = 40/104 (38%), Positives = 58/104 (55%)
Frame = -3
Query: 507 LSWQFNLQRVWEKIMHGKAVTPSLENDFKISSKETLPPHEFLYDNVSNGFPSHNYSARSA 328
+SWQF LQR+WE + G+A + S ++D K +E +S
Sbjct: 391 MSWQFGLQRIWESLARGEAKSNS-KSDSKGKEEE-----------------------KSF 426
Query: 327 VIPSLPLTIRWLRDCVKEHPSIRLQVLVTGSLHLVGDVLKLLKR 196
V SLP+ + WLRD ++ +R QVLVTGSLHLVGD+L+ +K+
Sbjct: 427 VFSSLPVAVDWLRDNARQSKQVRFQVLVTGSLHLVGDLLRFIKK 470
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 440,027,788
Number of Sequences: 1393205
Number of extensions: 8777924
Number of successful extensions: 20697
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 20239
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20680
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)