Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003391A_C01 KMC003391A_c01
(515 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
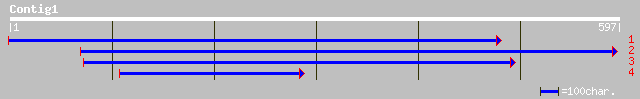
ref|NP_176564.1| putative U3 small nucleolar ribonucleoprotein p... 106 2e-24
ref|NP_593785.1| hypothetical protein [Schizosaccharomyces pombe... 84 1e-15
gb|EAA03875.1| agCP2306 [Anopheles gambiae str. PEST] 74 1e-14
gb|AAH37602.1| Unknown (protein for IMAGE:5135392) [Mus musculus] 75 2e-13
ref|NP_219484.1| hypothetical protein MGC19606 [Homo sapiens] gi... 75 2e-13
>ref|NP_176564.1| putative U3 small nucleolar ribonucleoprotein protein; protein id:
At1g63780.1, supported by cDNA: 418., supported by cDNA:
gi_20260205 [Arabidopsis thaliana]
gi|25347957|pir||H96662 hypothetical protein T12P18.20
[imported] - Arabidopsis thaliana
gi|12324948|gb|AAG52427.1|AC011622_15 putative U3 small
nucleolar ribonucleoprotein protein; 1537-3735
[Arabidopsis thaliana]
gi|12325007|gb|AAG52449.1|AC010852_6 putative U3 small
nucleolar ribonucleoprotein protein; 73469-75667
[Arabidopsis thaliana] gi|20260206|gb|AAM13001.1|
putative U3 small nucleolar ribonucleoprotein protein
[Arabidopsis thaliana] gi|21387041|gb|AAM47924.1|
putative U3 small nucleolar ribonucleoprotein protein
[Arabidopsis thaliana] gi|21593725|gb|AAM65692.1|
putative U3 small nucleolar ribonucleoprotein
[Arabidopsis thaliana]
Length = 294
Score = 106 bits (264), Expect(2) = 2e-24
Identities = 50/63 (79%), Positives = 58/63 (91%), Gaps = 1/63 (1%)
Frame = -3
Query: 513 IVTLTNQSDYISFRHHIYEK-PGGPKSIELKEIGPRFELRLYQIKLGTVDQAEAQTEWVI 337
IVT +NQSDYISFR+H+Y+K GGPKSIELKEIGPRFELRLYQ+KLGTV+Q EA+ EWVI
Sbjct: 220 IVTFSNQSDYISFRNHVYDKGEGGPKSIELKEIGPRFELRLYQVKLGTVEQNEAEIEWVI 279
Query: 336 RPF 328
RP+
Sbjct: 280 RPY 282
Score = 27.7 bits (60), Expect(2) = 2e-24
Identities = 10/14 (71%), Positives = 13/14 (92%)
Frame = -1
Query: 332 PFMNTSKKRKFLSD 291
P+MNTSKKRKF+ +
Sbjct: 281 PYMNTSKKRKFIGE 294
>ref|NP_593785.1| hypothetical protein [Schizosaccharomyces pombe]
gi|6226440|sp|O13823|YEE7_SCHPO Hypothetical protein
C19A8.07c in chromosome I gi|7490888|pir||T37954
hypothetical protein SPAC19A8.07c - fission yeast
(Schizosaccharomyces pombe) gi|2388906|emb|CAB11643.1|
hypothetical protein [Schizosaccharomyces pombe]
Length = 289
Score = 84.0 bits (206), Expect = 1e-15
Identities = 37/65 (56%), Positives = 46/65 (69%)
Frame = -3
Query: 513 IVTLTNQSDYISFRHHIYEKPGGPKSIELKEIGPRFELRLYQIKLGTVDQAEAQTEWVIR 334
+VT N DYISFRHHIY K G PK I L E GPRFE++L++I LGTVD +A EW ++
Sbjct: 219 VVTFANTDDYISFRHHIYAKTG-PKQIILSEAGPRFEMKLFEITLGTVDMVDADVEWKLK 277
Query: 333 PFHEH 319
P+ H
Sbjct: 278 PYQRH 282
>gb|EAA03875.1| agCP2306 [Anopheles gambiae str. PEST]
Length = 320
Score = 73.6 bits (179), Expect(2) = 1e-14
Identities = 35/63 (55%), Positives = 45/63 (70%), Gaps = 1/63 (1%)
Frame = -3
Query: 513 IVTLTNQSDYISFRHHIYEKPGGPKSIELKEIGPRFELRLYQIKLGTVDQAE-AQTEWVI 337
++T N D+ISFRHH + K +EL E GPRF+L+LYQIKLGTVD+ + A TEWV
Sbjct: 236 VMTFANHDDFISFRHHTFRMVD--KQLELTECGPRFQLKLYQIKLGTVDELDAADTEWVY 293
Query: 336 RPF 328
RP+
Sbjct: 294 RPY 296
Score = 26.9 bits (58), Expect(2) = 1e-14
Identities = 10/14 (71%), Positives = 13/14 (92%)
Frame = -1
Query: 332 PFMNTSKKRKFLSD 291
P+MNT+ KR+FLSD
Sbjct: 295 PYMNTAAKRRFLSD 308
>gb|AAH37602.1| Unknown (protein for IMAGE:5135392) [Mus musculus]
Length = 296
Score = 74.7 bits (182), Expect(2) = 2e-13
Identities = 34/63 (53%), Positives = 47/63 (73%), Gaps = 1/63 (1%)
Frame = -3
Query: 513 IVTLTNQSDYISFRHHIYEKPGGPKSIELKEIGPRFELRLYQIKLGTVDQ-AEAQTEWVI 337
++T NQ DYISFRHH+Y+K +++EL E+GPRFEL+LY I+LGT++Q A A EW
Sbjct: 222 VITFANQDDYISFRHHVYKKTDH-RNVELTEVGPRFELKLYMIRLGTLEQEATADVEWRW 280
Query: 336 RPF 328
P+
Sbjct: 281 HPY 283
Score = 21.9 bits (45), Expect(2) = 2e-13
Identities = 8/13 (61%), Positives = 11/13 (84%)
Frame = -1
Query: 332 PFMNTSKKRKFLS 294
P+ NT++KR FLS
Sbjct: 282 PYTNTARKRVFLS 294
>ref|NP_219484.1| hypothetical protein MGC19606 [Homo sapiens]
gi|14603153|gb|AAH10042.1|AAH10042 Unknown (protein for
MGC:19606) [Homo sapiens]
Length = 291
Score = 74.7 bits (182), Expect(2) = 2e-13
Identities = 34/63 (53%), Positives = 47/63 (73%), Gaps = 1/63 (1%)
Frame = -3
Query: 513 IVTLTNQSDYISFRHHIYEKPGGPKSIELKEIGPRFELRLYQIKLGTVDQ-AEAQTEWVI 337
++T NQ DYISFRHH+Y+K +++EL E+GPRFEL+LY I+LGT++Q A A EW
Sbjct: 217 VITFANQDDYISFRHHVYKKTDH-RNVELTEVGPRFELKLYMIRLGTLEQEATADVEWRW 275
Query: 336 RPF 328
P+
Sbjct: 276 HPY 278
Score = 21.9 bits (45), Expect(2) = 2e-13
Identities = 8/13 (61%), Positives = 11/13 (84%)
Frame = -1
Query: 332 PFMNTSKKRKFLS 294
P+ NT++KR FLS
Sbjct: 277 PYTNTARKRVFLS 289
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 399,266,191
Number of Sequences: 1393205
Number of extensions: 7525993
Number of successful extensions: 11652
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 11487
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11620
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16154357632
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)