Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003388A_C01 KMC003388A_c01
(570 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
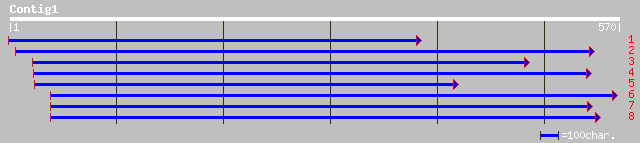
ref|NP_082323.1| RIKEN cDNA 1500002O20; expressed sequence N2809... 48 1e-04
emb|CAB66541.1| hypothetical protein [Homo sapiens] gi|14250798|... 47 1e-04
ref|NP_061981.1| hypothetical protein FLJ12886 [Homo sapiens] gi... 47 1e-04
dbj|BAC30328.1| unnamed protein product [Mus musculus] 45 6e-04
ref|XP_138185.1| similar to Amphoterin [Mus musculus] 35 0.66
>ref|NP_082323.1| RIKEN cDNA 1500002O20; expressed sequence N28092 [Mus musculus]
gi|12836835|dbj|BAB23829.1| unnamed protein product [Mus
musculus] gi|21707691|gb|AAH34211.1| RIKEN cDNA
1500002O20 gene [Mus musculus]
Length = 520
Score = 47.8 bits (112), Expect = 1e-04
Identities = 31/99 (31%), Positives = 50/99 (50%), Gaps = 20/99 (20%)
Frame = -3
Query: 445 MDSNVPSLFAIPF---KKKDENPRAQ----------------HGSYISALWKLRDQILSM 323
+D+ V +LF +PF + ++ENP H S+ S + KLR Q++SM
Sbjct: 422 LDAEV-NLFLVPFMDSEAENENPPRAGPGSSPLFSLLPGYRGHPSFQSLVSKLRSQVMSM 480
Query: 322 KSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 209
P + + +E+ W +A+IW+ VK S + EY R L
Sbjct: 481 ARPQLSHTILTEKNWFHYAARIWDGVKKSSALAEYSRLL 519
>emb|CAB66541.1| hypothetical protein [Homo sapiens]
gi|14250798|gb|AAH08869.1|AAH08869 hypothetical protein
F17127_1 [Homo sapiens]
Length = 520
Score = 47.4 bits (111), Expect = 1e-04
Identities = 31/99 (31%), Positives = 49/99 (49%), Gaps = 20/99 (20%)
Frame = -3
Query: 445 MDSNVPSLFAIPF---KKKDENPRAQ----------------HGSYISALWKLRDQILSM 323
+DS V +LF +PF + + ENP H S+ S + KLR Q++SM
Sbjct: 422 LDSEV-NLFLVPFMDSEAESENPPRAGPGSSPLFSLLPGYRGHPSFQSLVSKLRSQVMSM 480
Query: 322 KSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 209
P + + +E+ W +A+IW+ V+ S + EY R L
Sbjct: 481 ARPQLSHTILTEKNWFHYAARIWDGVRKSSALAEYSRLL 519
>ref|NP_061981.1| hypothetical protein FLJ12886 [Homo sapiens]
gi|3169705|gb|AAC17932.1| F17127_1 [Homo sapiens]
Length = 528
Score = 47.4 bits (111), Expect = 1e-04
Identities = 31/99 (31%), Positives = 49/99 (49%), Gaps = 20/99 (20%)
Frame = -3
Query: 445 MDSNVPSLFAIPF---KKKDENPRAQ----------------HGSYISALWKLRDQILSM 323
+DS V +LF +PF + + ENP H S+ S + KLR Q++SM
Sbjct: 430 LDSEV-NLFLVPFMDSEAESENPPRAGPGSSPLFSLLPGYRGHPSFQSLVSKLRSQVMSM 488
Query: 322 KSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 209
P + + +E+ W +A+IW+ V+ S + EY R L
Sbjct: 489 ARPQLSHTILTEKNWFHYAARIWDGVRKSSALAEYSRLL 527
>dbj|BAC30328.1| unnamed protein product [Mus musculus]
Length = 520
Score = 45.1 bits (105), Expect = 6e-04
Identities = 30/99 (30%), Positives = 49/99 (49%), Gaps = 20/99 (20%)
Frame = -3
Query: 445 MDSNVPSLFAIPF---KKKDENPRAQ----------------HGSYISALWKLRDQILSM 323
+D+ V +LF +PF + ++ENP H S+ S + KLR Q++SM
Sbjct: 422 LDAEV-NLFLVPFMDSEAENENPPRAGPGSSPLFSLLPGYRGHPSFQSLVSKLRSQVMSM 480
Query: 322 KSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 209
P + + + + W +A+IW+ VK S + EY R L
Sbjct: 481 ARPQLSHTILTGKNWFHYAARIWDGVKKSSALAEYSRLL 519
>ref|XP_138185.1| similar to Amphoterin [Mus musculus]
Length = 416
Score = 35.0 bits (79), Expect = 0.66
Identities = 23/66 (34%), Positives = 32/66 (47%)
Frame = -3
Query: 496 EHKPEEHISSSMVHDSPMDSNVPSLFAIPFKKKDENPRAQHGSYISALWKLRDQILSMKS 317
+H+P SS ++ SP++ NVP L A PF +P LW+ +DQ KS
Sbjct: 257 QHQPS---SSQVLQQSPLERNVPILMAAPFPIHSSSP---------PLWEAKDQ---PKS 301
Query: 316 PSFTRP 299
TRP
Sbjct: 302 SPCTRP 307
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 478,858,035
Number of Sequences: 1393205
Number of extensions: 10002401
Number of successful extensions: 22026
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 21384
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22017
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)