Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
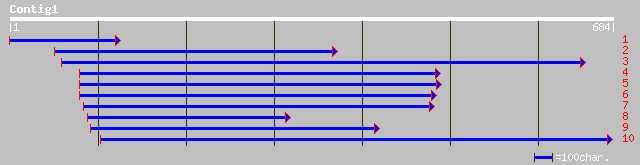
Query= KMC003380A_C01 KMC003380A_c01
(679 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_198835.1| calmodulin-binding protein; protein id: At5g401... 117 1e-25
dbj|BAB01134.1| gene_id:MMG15.17~unknown protein [Arabidopsis th... 105 7e-22
ref|NP_566834.1| calmodulin-binding protein; protein id: At3g281... 61 2e-08
ref|NP_650353.1| CG3259-PA [Drosophila melanogaster] gi|7299866|... 39 0.065
ref|XP_196822.1| hypothetical protein XP_196822 [Mus musculus] 38 0.15
>ref|NP_198835.1| calmodulin-binding protein; protein id: At5g40190.1 [Arabidopsis
thaliana] gi|10177506|dbj|BAB10900.1|
gene_id:MSN9.9~unknown protein [Arabidopsis thaliana]
Length = 183
Score = 117 bits (293), Expect = 1e-25
Identities = 52/82 (63%), Positives = 68/82 (82%)
Frame = -3
Query: 677 LHFQSQLCDAIRKEGVDVPDEYALGSWIPFCSVAHQVPKARMAEAFSLLRDLKLPVSGYA 498
L Q+QLCD ++KE VD+ +EY + SW+PFC VA VPK+RMAEAFS+LRDLKLPV+GY
Sbjct: 99 LQLQTQLCDMLKKESVDIGEEYRVDSWVPFCPVALDVPKSRMAEAFSVLRDLKLPVNGYG 158
Query: 497 VDVAVVQFSPVRELFSFVLGTT 432
+++ +V+FSPVRE+FSF LG T
Sbjct: 159 MEIGLVEFSPVREVFSFPLGNT 180
>dbj|BAB01134.1| gene_id:MMG15.17~unknown protein [Arabidopsis thaliana]
gi|26450294|dbj|BAC42263.1| unknown protein [Arabidopsis
thaliana]
Length = 184
Score = 105 bits (261), Expect = 7e-22
Identities = 47/80 (58%), Positives = 64/80 (79%)
Frame = -3
Query: 677 LHFQSQLCDAIRKEGVDVPDEYALGSWIPFCSVAHQVPKARMAEAFSLLRDLKLPVSGYA 498
+ Q QLC+ I+KEG ++ +EY + SW+PFCSVA VPK+R++E F +LRDLKL V GYA
Sbjct: 100 IQLQLQLCEVIKKEGFEIGEEYRVDSWVPFCSVAVDVPKSRISEGFLVLRDLKLLVYGYA 159
Query: 497 VDVAVVQFSPVRELFSFVLG 438
+D+ +V+FSPVRE+FSF LG
Sbjct: 160 MDIGLVEFSPVREVFSFGLG 179
>ref|NP_566834.1| calmodulin-binding protein; protein id: At3g28140.1, supported by
cDNA: 2194. [Arabidopsis thaliana]
Length = 168
Score = 60.8 bits (146), Expect = 2e-08
Identities = 24/46 (52%), Positives = 35/46 (75%)
Frame = -3
Query: 677 LHFQSQLCDAIRKEGVDVPDEYALGSWIPFCSVAHQVPKARMAEAF 540
+ Q QLC+ I+KEG ++ +EY + SW+PFCSVA VPK+R++E F
Sbjct: 100 IQLQLQLCEVIKKEGFEIGEEYRVDSWVPFCSVAVDVPKSRISEGF 145
>ref|NP_650353.1| CG3259-PA [Drosophila melanogaster] gi|7299866|gb|AAF55042.1|
CG3259-PA [Drosophila melanogaster]
Length = 588
Score = 38.9 bits (89), Expect = 0.065
Identities = 22/71 (30%), Positives = 37/71 (51%), Gaps = 2/71 (2%)
Frame = +2
Query: 467 PERIEQPRHPPRNQKPATSGPATEKTPPPSSPWEPDARRSKTESTNQARTH--PEHQHLP 640
P + E PP +K A P E P + P EP++R+S + S + +H PE +P
Sbjct: 265 PSKPEPEMVPPLKEKEAVVRPVQET--PTAPPAEPESRKSSSRSRRSSGSHRQPEPNPIP 322
Query: 641 SESRHRAATEN 673
+ES+ + A ++
Sbjct: 323 NESQSQPAVQS 333
>ref|XP_196822.1| hypothetical protein XP_196822 [Mus musculus]
Length = 265
Score = 37.7 bits (86), Expect = 0.15
Identities = 19/58 (32%), Positives = 24/58 (40%)
Frame = +2
Query: 470 ERIEQPRHPPRNQKPATSGPATEKTPPPSSPWEPDARRSKTESTNQARTHPEHQHLPS 643
+R P H P N+ P SG + P P P AR S + R HP H P+
Sbjct: 59 DRQPGPLHDPENRNPRLSGESPRSHPHSPRPKPPSARASGVAHRGKPRVHPLHARDPA 116
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 545,064,189
Number of Sequences: 1393205
Number of extensions: 11342546
Number of successful extensions: 59989
Number of sequences better than 10.0: 247
Number of HSP's better than 10.0 without gapping: 47463
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 57578
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29987172312
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)