Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003375A_C01 KMC003375A_c01
(558 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
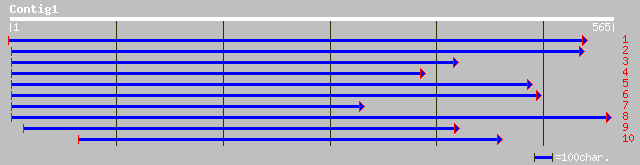
ref|NP_200887.1| ABC transporter family protein; protein id: At5... 99 4e-20
dbj|BAC55994.1| putative ABC transporter [Oryza sativa (japonica... 95 7e-19
gb|AAH46677.1| Similar to ATP-binding cassette, sub-family F (GC... 78 6e-14
ref|NP_573057.1| CG9281-PB [Drosophila melanogaster] gi|24642252... 77 1e-13
gb|AAL39441.1| GM14873p [Drosophila melanogaster] 77 1e-13
>ref|NP_200887.1| ABC transporter family protein; protein id: At5g60790.1
[Arabidopsis thaliana] gi|10176907|dbj|BAB10100.1| ABC
transporter [Arabidopsis thaliana]
gi|22655234|gb|AAM98207.1| ABC transporter homolog
PnATH-like protein [Arabidopsis thaliana]
gi|24030366|gb|AAN41346.1| putative ABC transporter
homolog PnATH [Arabidopsis thaliana]
Length = 595
Score = 99.0 bits (245), Expect = 4e-20
Identities = 44/53 (83%), Positives = 46/53 (86%)
Frame = -2
Query: 557 EWDGVLLLVSHDFRLINQVAHEIWVCENQAVKKWEGGIMDFKRHLKAKAGLSD 399
EWDG L+LVSHDFRLINQVAHEIWVCE Q + KW G IMDFKRHLKAKAGL D
Sbjct: 543 EWDGGLVLVSHDFRLINQVAHEIWVCEKQCITKWNGDIMDFKRHLKAKAGLED 595
>dbj|BAC55994.1| putative ABC transporter [Oryza sativa (japonica cultivar-group)]
Length = 592
Score = 94.7 bits (234), Expect = 7e-19
Identities = 42/53 (79%), Positives = 47/53 (88%)
Frame = -2
Query: 557 EWDGVLLLVSHDFRLINQVAHEIWVCENQAVKKWEGGIMDFKRHLKAKAGLSD 399
EWDG L+LVSHDFRLINQVA EIWVCE QAV +WEG IMDFK HL+++AGLSD
Sbjct: 540 EWDGGLVLVSHDFRLINQVAQEIWVCEKQAVTRWEGDIMDFKEHLRSRAGLSD 592
>gb|AAH46677.1| Similar to ATP-binding cassette, sub-family F (GCN20), member 2
[Xenopus laevis]
Length = 607
Score = 78.2 bits (191), Expect = 6e-14
Identities = 33/50 (66%), Positives = 42/50 (84%)
Frame = -2
Query: 557 EWDGVLLLVSHDFRLINQVAHEIWVCENQAVKKWEGGIMDFKRHLKAKAG 408
E++G ++LVSHDFRLI QVA EIWVCE QAV KW GGI+ +K+HLK++ G
Sbjct: 556 EFEGGMMLVSHDFRLIQQVAQEIWVCEKQAVTKWSGGILSYKQHLKSRLG 605
>ref|NP_573057.1| CG9281-PB [Drosophila melanogaster] gi|24642252|ref|NP_727881.1|
CG9281-PC [Drosophila melanogaster]
gi|7293109|gb|AAF48493.1| CG9281-PB [Drosophila
melanogaster] gi|16768776|gb|AAL28607.1| LD02975p
[Drosophila melanogaster] gi|22832283|gb|AAN09361.1|
CG9281-PC [Drosophila melanogaster]
Length = 611
Score = 77.4 bits (189), Expect = 1e-13
Identities = 33/48 (68%), Positives = 41/48 (84%)
Frame = -2
Query: 557 EWDGVLLLVSHDFRLINQVAHEIWVCENQAVKKWEGGIMDFKRHLKAK 414
++DG ++LVSHDFRLINQVA EIWVCE + V KW+GGI+D+K HLK K
Sbjct: 551 DFDGGMVLVSHDFRLINQVAEEIWVCEKETVTKWKGGILDYKDHLKNK 598
>gb|AAL39441.1| GM14873p [Drosophila melanogaster]
Length = 304
Score = 77.4 bits (189), Expect = 1e-13
Identities = 33/48 (68%), Positives = 41/48 (84%)
Frame = -2
Query: 557 EWDGVLLLVSHDFRLINQVAHEIWVCENQAVKKWEGGIMDFKRHLKAK 414
++DG ++LVSHDFRLINQVA EIWVCE + V KW+GGI+D+K HLK K
Sbjct: 244 DFDGGMVLVSHDFRLINQVAEEIWVCEKETVTKWKGGILDYKDHLKNK 291
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 486,810,450
Number of Sequences: 1393205
Number of extensions: 10376284
Number of successful extensions: 25835
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 24285
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25521
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)