Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003373A_C02 KMC003373A_c02
(603 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
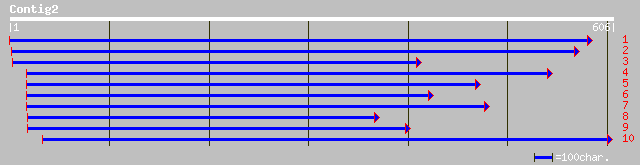
ref|NP_175587.1| IAA-Ala hydrolase (IAR3); protein id: At1g51760... 124 7e-28
dbj|BAB85405.1| putative IAA-Ala hydrolase [Oryza sativa (japoni... 120 2e-26
gb|AAO25632.1| IAA-amino acid hydrolase [Oryza sativa (indica cu... 120 2e-26
ref|NP_175589.1| auxin conjugate hydrolase (ILL5); protein id: A... 114 1e-24
emb|CAA73905.1| JR3 protein [Arabidopsis thaliana] 112 4e-24
>ref|NP_175587.1| IAA-Ala hydrolase (IAR3); protein id: At1g51760.1, supported by
cDNA: gi_14030706 [Arabidopsis thaliana]
gi|25319488|pir||F96556 IAA-Ala hydrolase (IAR3)
[imported] - Arabidopsis thaliana
gi|3421384|gb|AAC32192.1| IAA-Ala hydrolase; IAA-amino
acid hydrolase [Arabidopsis thaliana]
gi|12321681|gb|AAG50883.1|AC025294_21 IAA-Ala hydrolase
(IAR3) [Arabidopsis thaliana]
gi|14030707|gb|AAK53028.1|AF375444_1 At1g51760/F19C24_4
[Arabidopsis thaliana] gi|23506081|gb|AAN28900.1|
At1g51760/F19C24_4 [Arabidopsis thaliana]
Length = 440
Score = 124 bits (312), Expect = 7e-28
Identities = 62/133 (46%), Positives = 90/133 (67%)
Frame = -3
Query: 595 QAAVHRCNATVNFFEEVSPLYPPTINDAGLHEQFRDVALNLLGADKVHFDLPPVTASEDF 416
QA+V+ CNATV+F EE P +PPT+ND LH+ F++V+ ++LG + + ++ P+ SEDF
Sbjct: 311 QASVNMCNATVDFIEEEKPFFPPTVNDKALHQFFKNVSGDMLGIEN-YVEMQPLMGSEDF 369
Query: 415 SFYQEVMPGYFFFLGMHKPSNDPRAHILHSPHLFINEEGLPYGAALHASLAVNYLQKYQQ 236
SFYQ+ +PG+F F+GM + P A HSP+ +NEE LPYGA+LHAS+A YL + +
Sbjct: 370 SFYQQAIPGHFSFVGMQNKARSPMAS-PHSPYFEVNEELLPYGASLHASMATRYLLELKA 428
Query: 235 DGSTVEGKYRDEL 197
K +DEL
Sbjct: 429 STLNKSNK-KDEL 440
>dbj|BAB85405.1| putative IAA-Ala hydrolase [Oryza sativa (japonica cultivar-group)]
Length = 442
Score = 120 bits (300), Expect = 2e-26
Identities = 64/136 (47%), Positives = 89/136 (65%), Gaps = 1/136 (0%)
Frame = -3
Query: 601 IGQAAVHRCNATVNFFEEVSPLYPPTINDAGLHEQFRDVALNLLGADKVHFDLPPVTASE 422
+ QA+V RCNA V+F ++ P +PPTIN AGLH+ F VA ++G V D P+ +E
Sbjct: 310 VSQASVQRCNAVVDFLDKDRPFFPPTINSAGLHDFFVKVASEMVGPKNVR-DKQPLMGAE 368
Query: 421 DFSFYQEVMPG-YFFFLGMHKPSNDPRAHILHSPHLFINEEGLPYGAALHASLAVNYLQK 245
DF+FY + +P Y++FLGM+ + P+A HSP+ INE+ LPYGAAL ASLA YL +
Sbjct: 369 DFAFYADAIPATYYYFLGMYNETRGPQAP-HHSPYFTINEDALPYGAALQASLAARYLLE 427
Query: 244 YQQDGSTVEGKYRDEL 197
+Q +T + K DEL
Sbjct: 428 HQPP-TTGKAKAHDEL 442
>gb|AAO25632.1| IAA-amino acid hydrolase [Oryza sativa (indica cultivar-group)]
Length = 442
Score = 120 bits (300), Expect = 2e-26
Identities = 64/136 (47%), Positives = 89/136 (65%), Gaps = 1/136 (0%)
Frame = -3
Query: 601 IGQAAVHRCNATVNFFEEVSPLYPPTINDAGLHEQFRDVALNLLGADKVHFDLPPVTASE 422
+ QA+V RCNA V+F ++ P +PPTIN AGLH+ F VA ++G V D P+ +E
Sbjct: 310 VSQASVQRCNAVVDFLDKDRPFFPPTINSAGLHDFFVKVASEMVGPKNVR-DKQPLMGAE 368
Query: 421 DFSFYQEVMPG-YFFFLGMHKPSNDPRAHILHSPHLFINEEGLPYGAALHASLAVNYLQK 245
DF+FY + +P Y++FLGM+ + P+A HSP+ INE+ LPYGAAL ASLA YL +
Sbjct: 369 DFAFYADAIPATYYYFLGMYNETRGPQAP-HHSPYFTINEDALPYGAALQASLATRYLLE 427
Query: 244 YQQDGSTVEGKYRDEL 197
+Q +T + K DEL
Sbjct: 428 HQPP-TTGKAKAHDEL 442
>ref|NP_175589.1| auxin conjugate hydrolase (ILL5); protein id: At1g51780.1
[Arabidopsis thaliana] gi|25319490|pir||H96556 auxin
conjugate hydrolase (ILL5) [imported] - Arabidopsis
thaliana gi|5725649|gb|AAD48152.1| auxin conjugate
hydrolase [Arabidopsis thaliana]
gi|12321667|gb|AAG50869.1|AC025294_7 auxin conjugate
hydrolase (ILL5) [Arabidopsis thaliana]
Length = 435
Score = 114 bits (284), Expect = 1e-24
Identities = 56/115 (48%), Positives = 83/115 (71%)
Frame = -3
Query: 595 QAAVHRCNATVNFFEEVSPLYPPTINDAGLHEQFRDVALNLLGADKVHFDLPPVTASEDF 416
QA+V+ CNATV+F E+ +P +PPT+N+ LH +++V++++LG + + + PV SEDF
Sbjct: 311 QASVNMCNATVDFLEDETPPFPPTVNNKTLHLFYKNVSVDMLGIEN-YVETLPVMVSEDF 369
Query: 415 SFYQEVMPGYFFFLGMHKPSNDPRAHILHSPHLFINEEGLPYGAALHASLAVNYL 251
+FYQ+ +PG+F F+GM S+ P A+ HSP +NEE LPYGA+L ASLA YL
Sbjct: 370 AFYQQAIPGHFSFVGMQNKSHSPMAN-PHSPFFEVNEELLPYGASLLASLATRYL 423
>emb|CAA73905.1| JR3 protein [Arabidopsis thaliana]
Length = 444
Score = 112 bits (280), Expect = 4e-24
Identities = 59/134 (44%), Positives = 87/134 (64%), Gaps = 1/134 (0%)
Frame = -3
Query: 595 QAAVHRCNATVNFFEEVSPLY-PPTINDAGLHEQFRDVALNLLGADKVHFDLPPVTASED 419
QA+V+ CNATV+F + PPT+ND LH+ F++V+ ++LG + + ++ P+ SED
Sbjct: 314 QASVNMCNATVDFIARGETFFXPPTVNDKALHQFFKNVSGDMLGIEN-YVEMQPLMGSED 372
Query: 418 FSFYQEVMPGYFFFLGMHKPSNDPRAHILHSPHLFINEEGLPYGAALHASLAVNYLQKYQ 239
FSFYQ+ +PG+F F+GM + P A HSP+ +NEE LPYGA+LHAS+A YL + +
Sbjct: 373 FSFYQQAIPGHFSFVGMQNKARSPMAS-PHSPYFEVNEELLPYGASLHASMATRYLLELK 431
Query: 238 QDGSTVEGKYRDEL 197
K +DEL
Sbjct: 432 ASTLNKSNK-KDEL 444
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 488,546,332
Number of Sequences: 1393205
Number of extensions: 9959019
Number of successful extensions: 19857
Number of sequences better than 10.0: 177
Number of HSP's better than 10.0 without gapping: 19322
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19716
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23711793746
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)