Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003369A_C01 KMC003369A_c01
(931 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
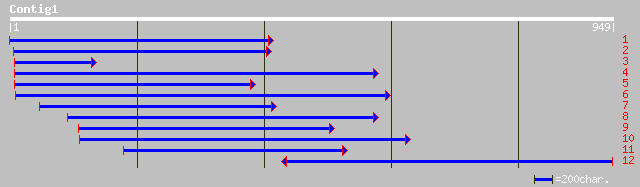
dbj|BAB21610.2| mangrin [Bruguiera sexangula] 310 2e-83
emb|CAB95731.1| allene oxide cyclase [Lycopersicon esculentum] g... 300 2e-80
emb|CAD38519.1| allene oxide cyclase [Oryza sativa (japonica cul... 296 2e-79
pir||S57813 hypothetical protein (clone TPP15) - tomato (fragmen... 295 7e-79
gb|AAN37418.1| allene oxide cyclase [Solanum tuberosum] 291 1e-77
>dbj|BAB21610.2| mangrin [Bruguiera sexangula]
Length = 256
Score = 310 bits (795), Expect = 2e-83
Identities = 158/253 (62%), Positives = 189/253 (74%), Gaps = 3/253 (1%)
Frame = -3
Query: 929 LKMISSSLKLSNPSSIPPNQTQNQVVSSLSQSFP---TKSFKISASPQVPSTKRSSKNTT 759
L+ +SSS+K+ P+ Q V + P T+S S+ + S +
Sbjct: 8 LRTVSSSVKVVGPARSKSATVPTQTVLPFKFTNPSLLTRSLSFSSK----GSSFDSFSVP 63
Query: 758 TTAFFFSNNSKQDHDSSRPAKVQELSVYEINERDRGSPAYLRLSQKPVNSLGDLVPFSNK 579
+F + + D+SRP KVQEL VYE+NERDRGSPA LRLSQKPVNSLGDLVPFSNK
Sbjct: 64 KRSFSCRSQATPSDDASRPTKVQELCVYEMNERDRGSPAVLRLSQKPVNSLGDLVPFSNK 123
Query: 578 LYSGNLEKRLGITTGLCVLIQNVPEKKGDRYEAIYSFYFGDYGHISVQGAYLTYQDTYLA 399
+YSG+L+KR+G+T G+C+LIQN PEKKGDR+EAIYSFYFG YGHI+VQGAYLTY+DT+LA
Sbjct: 124 VYSGDLQKRIGVTAGICILIQNKPEKKGDRFEAIYSFYFGGYGHIAVQGAYLTYEDTHLA 183
Query: 398 VTGGSGIFEGVYGQVKLQQLVFPFKLFYTFYLKGIPDLPAELLGKPVDPSPDVEPSPAAK 219
VTGGSGIFEGV GQVKLQQLV+PFKLFYTFYL+GI DLP EL KPV+P P VEP PAAK
Sbjct: 184 VTGGSGIFEGVSGQVKLQQLVYPFKLFYTFYLRGIKDLPEELTKKPVEPHPSVEPMPAAK 243
Query: 218 ATEPHATLPNFTN 180
A EPHA + NFT+
Sbjct: 244 ACEPHAVVANFTD 256
>emb|CAB95731.1| allene oxide cyclase [Lycopersicon esculentum]
gi|14423351|gb|AAK62358.1|AF384374_1 allene oxide cylase
[Lycopersicon esculentum]
Length = 244
Score = 300 bits (768), Expect = 2e-80
Identities = 154/227 (67%), Positives = 179/227 (78%), Gaps = 3/227 (1%)
Frame = -3
Query: 851 SSLSQSFPTK---SFKISASPQVPSTKRSSKNTTTTAFFFSNNSKQDHDSSRPAKVQELS 681
S LS +F TK SFK+ +P + + + +TT + FS S+ S +VQELS
Sbjct: 19 SKLSSAFQTKKIQSFKLP-NPLISQNHKLTTTSTTASRSFSCKSQSTSTDSTNTEVQELS 77
Query: 680 VYEINERDRGSPAYLRLSQKPVNSLGDLVPFSNKLYSGNLEKRLGITTGLCVLIQNVPEK 501
VYEINERDRGSPAYLRLSQK VNSLGDLVPFSNKLY+ +L+KR+GIT GLC+LI++ EK
Sbjct: 78 VYEINERDRGSPAYLRLSQKTVNSLGDLVPFSNKLYTADLKKRIGITAGLCILIKHEEEK 137
Query: 500 KGDRYEAIYSFYFGDYGHISVQGAYLTYQDTYLAVTGGSGIFEGVYGQVKLQQLVFPFKL 321
KGDRYEA+YSFYFGDYGHI+VQGAYLTY++TYLAVTGGSGIF GV GQVKLQQL+FPFKL
Sbjct: 138 KGDRYEAVYSFYFGDYGHIAVQGAYLTYEETYLAVTGGSGIFAGVSGQVKLQQLIFPFKL 197
Query: 320 FYTFYLKGIPDLPAELLGKPVDPSPDVEPSPAAKATEPHATLPNFTN 180
FYTFYLKGIP LP+ELL V PSP VEP+P AKA E A L N+TN
Sbjct: 198 FYTFYLKGIPGLPSELLCTAVPPSPTVEPTPEAKACEEGAALKNYTN 244
>emb|CAD38519.1| allene oxide cyclase [Oryza sativa (japonica cultivar-group)]
Length = 240
Score = 296 bits (759), Expect = 2e-79
Identities = 142/178 (79%), Positives = 160/178 (89%), Gaps = 1/178 (0%)
Frame = -3
Query: 710 SRPAKVQELSVYEINERDRGSPAYLRLSQKPV-NSLGDLVPFSNKLYSGNLEKRLGITTG 534
+RPAKVQE+ VYEINERDR SPAYLRLS K N+LGDLVPF+NKLYSG+L+KRLGI+ G
Sbjct: 63 ARPAKVQEMFVYEINERDRESPAYLRLSAKQTENALGDLVPFTNKLYSGSLDKRLGISAG 122
Query: 533 LCVLIQNVPEKKGDRYEAIYSFYFGDYGHISVQGAYLTYQDTYLAVTGGSGIFEGVYGQV 354
+C+LIQ+VPE+ GDRYEAIYSFYFGDYGHISVQG YLTY+++YLAVTGGSG+FEG YGQV
Sbjct: 123 ICILIQHVPERNGDRYEAIYSFYFGDYGHISVQGPYLTYEESYLAVTGGSGVFEGAYGQV 182
Query: 353 KLQQLVFPFKLFYTFYLKGIPDLPAELLGKPVDPSPDVEPSPAAKATEPHATLPNFTN 180
KL Q+VFPFK+FYTFYLKGIPDLP ELL PV PSP VEP+PAAKATEPHA L NFTN
Sbjct: 183 KLNQIVFPFKIFYTFYLKGIPDLPRELLCTPVPPSPTVEPTPAAKATEPHACLNNFTN 240
>pir||S57813 hypothetical protein (clone TPP15) - tomato (fragment)
gi|924632|gb|AAA80500.1| unknown
Length = 214
Score = 295 bits (755), Expect = 7e-79
Identities = 148/215 (68%), Positives = 173/215 (79%)
Frame = -3
Query: 824 KSFKISASPQVPSTKRSSKNTTTTAFFFSNNSKQDHDSSRPAKVQELSVYEINERDRGSP 645
+SFK+ +P + + + +TT + FS S+ S +VQELSVYEINERDRGSP
Sbjct: 1 QSFKLP-NPLISQNHKLTTTSTTASRSFSCKSQSTSTDSTNTEVQELSVYEINERDRGSP 59
Query: 644 AYLRLSQKPVNSLGDLVPFSNKLYSGNLEKRLGITTGLCVLIQNVPEKKGDRYEAIYSFY 465
AYLRLSQK VNSLGDLVPFSNKLY+ +L+KR+GIT GLC+LI++ EKKGDRYEA+YSFY
Sbjct: 60 AYLRLSQKTVNSLGDLVPFSNKLYTADLKKRIGITAGLCILIKHEEEKKGDRYEAVYSFY 119
Query: 464 FGDYGHISVQGAYLTYQDTYLAVTGGSGIFEGVYGQVKLQQLVFPFKLFYTFYLKGIPDL 285
FGDYGHI+VQGAYLTY++TYLAVTGGSGIF GV GQVKLQQL+FPFKLFYTFYLKGIP L
Sbjct: 120 FGDYGHIAVQGAYLTYEETYLAVTGGSGIFAGVSGQVKLQQLIFPFKLFYTFYLKGIPGL 179
Query: 284 PAELLGKPVDPSPDVEPSPAAKATEPHATLPNFTN 180
P+ELL V PSP VEP+P AKA E A L N+TN
Sbjct: 180 PSELLCTAVPPSPTVEPTPEAKACEEGAALKNYTN 214
>gb|AAN37418.1| allene oxide cyclase [Solanum tuberosum]
Length = 246
Score = 291 bits (745), Expect = 1e-77
Identities = 148/223 (66%), Positives = 177/223 (79%)
Frame = -3
Query: 854 VSSLSQSFPTKSFKISASPQVPSTKRSSKNTTTTAFFFSNNSKQDHDSSRPAKVQELSVY 675
++SLSQ +S K+ +P + T + + +TT + FS S+ S +VQELSVY
Sbjct: 27 ITSLSQKI--QSLKLP-NPLISQTHKLTTTSTTASRSFSCKSQNSSTDSTNTEVQELSVY 83
Query: 674 EINERDRGSPAYLRLSQKPVNSLGDLVPFSNKLYSGNLEKRLGITTGLCVLIQNVPEKKG 495
E+NERDRGSPAYLRLSQK VNSLGDLVPFSNKLY+ +L+KR+GIT GLC+LI++ EKKG
Sbjct: 84 ELNERDRGSPAYLRLSQKNVNSLGDLVPFSNKLYTADLKKRIGITAGLCILIKHEEEKKG 143
Query: 494 DRYEAIYSFYFGDYGHISVQGAYLTYQDTYLAVTGGSGIFEGVYGQVKLQQLVFPFKLFY 315
DRYEA+YSFYFGDYG I+VQG+YLTY+D+YLAVTGGSGIF GV GQVKLQQL+FPFKLFY
Sbjct: 144 DRYEAVYSFYFGDYGQIAVQGSYLTYEDSYLAVTGGSGIFAGVSGQVKLQQLIFPFKLFY 203
Query: 314 TFYLKGIPDLPAELLGKPVDPSPDVEPSPAAKATEPHATLPNF 186
TFYLKGIPDLP+ELL V PSP VEP+P AKA E A L N+
Sbjct: 204 TFYLKGIPDLPSELLCTAVPPSPTVEPTPEAKACEDGAALKNY 246
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 796,970,897
Number of Sequences: 1393205
Number of extensions: 17868511
Number of successful extensions: 62477
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 55352
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 62087
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 51582455952
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)