Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003363A_C01 KMC003363A_c01
(561 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
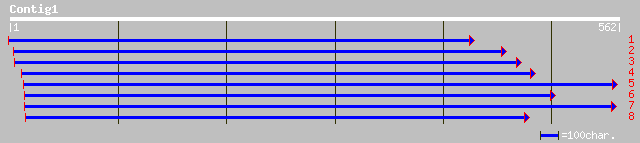
pir||T09598 cyclin 4, D-type - alfalfa gi|1150932|emb|CAA61334.1... 72 5e-12
emb|CAB40540.1| cyclin D3 [Medicago sativa] 72 5e-12
dbj|BAA33153.1| cyclin D [Pisum sativum] 71 8e-12
ref|NP_190576.1| Cyclin D3;3; protein id: At3g50070.1, supported... 57 2e-07
emb|CAA09854.1| cyclin D3.2 protein [Nicotiana tabacum] 51 1e-05
>pir||T09598 cyclin 4, D-type - alfalfa gi|1150932|emb|CAA61334.1| cyclin
[Medicago sativa]
Length = 386
Score = 72.0 bits (175), Expect = 5e-12
Identities = 37/59 (62%), Positives = 48/59 (80%), Gaps = 3/59 (5%)
Frame = -3
Query: 559 GVIDAAFNSDGSNDSWTVGSSLFSSSGPESPLFKKSRTQ---MKLSPLNRVIVGIVSTS 392
GVIDA F+SDGSNDSWTVG+S +S+S P+FKK++ Q M LSP+NRVIVGI++T+
Sbjct: 328 GVIDAVFSSDGSNDSWTVGASSYSTS---EPVFKKTKNQGQNMNLSPINRVIVGILATA 383
>emb|CAB40540.1| cyclin D3 [Medicago sativa]
Length = 378
Score = 72.0 bits (175), Expect = 5e-12
Identities = 37/59 (62%), Positives = 48/59 (80%), Gaps = 3/59 (5%)
Frame = -3
Query: 559 GVIDAAFNSDGSNDSWTVGSSLFSSSGPESPLFKKSRTQ---MKLSPLNRVIVGIVSTS 392
GVIDA F+SDGSNDSWTVG+S +S+S P+FKK++ Q M LSP+NRVIVGI++T+
Sbjct: 320 GVIDAVFSSDGSNDSWTVGASSYSTS---EPVFKKTKNQGQNMNLSPINRVIVGILATA 375
>dbj|BAA33153.1| cyclin D [Pisum sativum]
Length = 384
Score = 71.2 bits (173), Expect = 8e-12
Identities = 38/59 (64%), Positives = 47/59 (79%), Gaps = 3/59 (5%)
Frame = -3
Query: 559 GVIDAAFNSDGSNDSWTVGSSLFSSSGPESPLFKKSRTQ---MKLSPLNRVIVGIVSTS 392
GVIDA F+SDGSNDSW VGSS +S+S P+FKK++TQ LSPLNRVIVGI++T+
Sbjct: 323 GVIDAVFSSDGSNDSWKVGSSSYSTS---EPVFKKTKTQGQNRNLSPLNRVIVGILATA 378
>ref|NP_190576.1| Cyclin D3;3; protein id: At3g50070.1, supported by cDNA: 36056.,
supported by cDNA: gi_15450594, supported by cDNA:
gi_17380631 [Arabidopsis thaliana]
gi|11357237|pir||T45860 cyclin D3-like protein -
Arabidopsis thaliana gi|6522928|emb|CAB62115.1| cyclin
D3-like protein [Arabidopsis thaliana]
gi|15450595|gb|AAK96569.1| AT3g50070/F3A4_150
[Arabidopsis thaliana] gi|17380632|gb|AAL36079.1|
AT3g50070/F3A4_150 [Arabidopsis thaliana]
gi|21593092|gb|AAM65041.1| cyclin D3-like protein
[Arabidopsis thaliana]
Length = 361
Score = 56.6 bits (135), Expect = 2e-07
Identities = 30/61 (49%), Positives = 41/61 (67%), Gaps = 4/61 (6%)
Frame = -3
Query: 559 GVIDAAFNSDGSNDSWTVGSSLFSSSGPES-PLFKKSRT---QMKLSPLNRVIVGIVSTS 392
GV DA+F+SD SN+SW V +S SS P S PL K+ R QM+LS +NR+ ++S+S
Sbjct: 300 GVFDASFSSDSSNESWVVSASASVSSSPSSEPLLKRRRVQEQQMRLSSINRMFFDVLSSS 359
Query: 391 P 389
P
Sbjct: 360 P 360
>emb|CAA09854.1| cyclin D3.2 protein [Nicotiana tabacum]
Length = 367
Score = 50.8 bits (120), Expect = 1e-05
Identities = 28/57 (49%), Positives = 36/57 (63%)
Frame = -3
Query: 559 GVIDAAFNSDGSNDSWTVGSSLFSSSGPESPLFKKSRTQMKLSPLNRVIVGIVSTSP 389
GVIDA F+ D SNDSW+V SS+ SS P+ K M L+PL+ V V +V +SP
Sbjct: 311 GVIDAYFSCDSSNDSWSVASSISSSPEPQYKRIKTQDQTMTLAPLSSVSV-VVGSSP 366
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 488,271,065
Number of Sequences: 1393205
Number of extensions: 10269433
Number of successful extensions: 29541
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 28261
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29519
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)