Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003357A_C01 KMC003357A_c01
(668 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
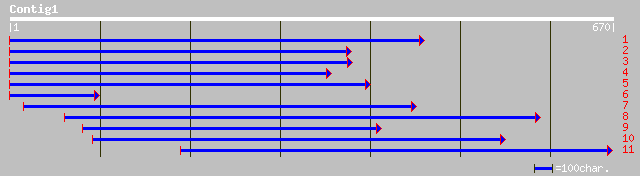
dbj|BAB18313.1| putative WRKY DNA binding protein [Oryza sativa ... 127 2e-28
gb|AAD38283.1|AC007789_9 putative WRKY DNA binding protein [Oryz... 127 2e-28
ref|NP_567644.1| WRKY family transcription factor; protein id: A... 98 9e-20
pir||T49114 hypothetical protein AT4g22070 - Arabidopsis thalian... 98 9e-20
ref|NP_192354.1| WRKY family transcription factor; protein id: A... 89 4e-17
>dbj|BAB18313.1| putative WRKY DNA binding protein [Oryza sativa (japonica
cultivar-group)] gi|13486842|dbj|BAB40073.1| putative
WRKY DNA binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 635
Score = 127 bits (318), Expect = 2e-28
Identities = 83/159 (52%), Positives = 95/159 (59%), Gaps = 17/159 (10%)
Frame = -3
Query: 666 SAPFPTVTLDLTHNPN------PLQFSRPQHSAP-FQIPQNFMSGPASFAQAAP---LYN 517
SAPFPTVTLDLTH P PL +RP AP FQ+P G + A A P LYN
Sbjct: 467 SAPFPTVTLDLTHAPPGAPNAVPLNAARPGAPAPQFQVP--LPGGGMAPAFAVPPQVLYN 524
Query: 516 QSKFSGLQLSSQQEVGSSHQLASQQPPQQQPS-------LADTVSAATAAITADPNFTAV 358
QSKFSGLQ+SS ++ A+ Q Q +P L+DTVSAA AAITADPNFT
Sbjct: 525 QSKFSGLQMSSDSAEAAAAAAAAAQFAQPRPPIGQLPGPLSDTVSAAAAAITADPNFTVA 584
Query: 357 LAAAISSIIGGSHANNNGSHNNNNNNNNNKSTMSNFSGN 241
LAAAI+SIIGG HA G+ N NN N N S +N S N
Sbjct: 585 LAAAITSIIGGQHAAAAGNSNANNTNTNTTSNTNNTSSN 623
>gb|AAD38283.1|AC007789_9 putative WRKY DNA binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 470
Score = 127 bits (318), Expect = 2e-28
Identities = 83/159 (52%), Positives = 95/159 (59%), Gaps = 17/159 (10%)
Frame = -3
Query: 666 SAPFPTVTLDLTHNPN------PLQFSRPQHSAP-FQIPQNFMSGPASFAQAAP---LYN 517
SAPFPTVTLDLTH P PL +RP AP FQ+P G + A A P LYN
Sbjct: 302 SAPFPTVTLDLTHAPPGAPNAVPLNAARPGAPAPQFQVP--LPGGGMAPAFAVPPQVLYN 359
Query: 516 QSKFSGLQLSSQQEVGSSHQLASQQPPQQQPS-------LADTVSAATAAITADPNFTAV 358
QSKFSGLQ+SS ++ A+ Q Q +P L+DTVSAA AAITADPNFT
Sbjct: 360 QSKFSGLQMSSDSAEAAAAAAAAAQFAQPRPPIGQLPGPLSDTVSAAAAAITADPNFTVA 419
Query: 357 LAAAISSIIGGSHANNNGSHNNNNNNNNNKSTMSNFSGN 241
LAAAI+SIIGG HA G+ N NN N N S +N S N
Sbjct: 420 LAAAITSIIGGQHAAAAGNSNANNTNTNTTSNTNNTSSN 458
>ref|NP_567644.1| WRKY family transcription factor; protein id: At4g22070.1,
supported by cDNA: gi_15990589 [Arabidopsis thaliana]
gi|20978775|sp|Q93WT0|WR31_ARATH Probable WRKY
transcription factor 31 (WRKY DNA-binding protein 31)
gi|15990590|gb|AAL11009.1| WRKY transcription factor 31
[Arabidopsis thaliana]
Length = 538
Score = 98.2 bits (243), Expect = 9e-20
Identities = 67/145 (46%), Positives = 86/145 (59%), Gaps = 5/145 (3%)
Frame = -3
Query: 666 SAPFPTVTLDLTHNPNPLQFSRPQHSAPFQIPQNFMSGPASFAQAA--PLYN---QSKFS 502
SAPFPT+TLDLT++PN + ++ Q Q PA Q +YN QSKFS
Sbjct: 409 SAPFPTITLDLTNSPNGNNPNMTTNNPLMQFAQRPGFNPAVLPQVVGQAMYNNQQQSKFS 468
Query: 501 GLQLSSQQEVGSSHQLASQQPPQQQPSLADTVSAATAAITADPNFTAVLAAAISSIIGGS 322
GLQL +Q Q+A+ S+A++VSAA+AAI +DPNF A LAAAI+SI+ GS
Sbjct: 469 GLQLPAQPL-----QIAATS------SVAESVSAASAAIASDPNFAAALAAAITSIMNGS 517
Query: 321 HANNNGSHNNNNNNNNNKSTMSNFS 247
SH NNN NNNN +T +N S
Sbjct: 518 ------SHQNNNTNNNNVATSNNDS 536
>pir||T49114 hypothetical protein AT4g22070 - Arabidopsis thaliana
gi|2961352|emb|CAA18110.1| putative protein [Arabidopsis
thaliana] gi|7269052|emb|CAB79162.1| putative protein
[Arabidopsis thaliana]
Length = 458
Score = 98.2 bits (243), Expect = 9e-20
Identities = 67/145 (46%), Positives = 86/145 (59%), Gaps = 5/145 (3%)
Frame = -3
Query: 666 SAPFPTVTLDLTHNPNPLQFSRPQHSAPFQIPQNFMSGPASFAQAA--PLYN---QSKFS 502
SAPFPT+TLDLT++PN + ++ Q Q PA Q +YN QSKFS
Sbjct: 329 SAPFPTITLDLTNSPNGNNPNMTTNNPLMQFAQRPGFNPAVLPQVVGQAMYNNQQQSKFS 388
Query: 501 GLQLSSQQEVGSSHQLASQQPPQQQPSLADTVSAATAAITADPNFTAVLAAAISSIIGGS 322
GLQL +Q Q+A+ S+A++VSAA+AAI +DPNF A LAAAI+SI+ GS
Sbjct: 389 GLQLPAQPL-----QIAATS------SVAESVSAASAAIASDPNFAAALAAAITSIMNGS 437
Query: 321 HANNNGSHNNNNNNNNNKSTMSNFS 247
SH NNN NNNN +T +N S
Sbjct: 438 ------SHQNNNTNNNNVATSNNDS 456
>ref|NP_192354.1| WRKY family transcription factor; protein id: At4g04450.1,
supported by cDNA: gi_15990593 [Arabidopsis thaliana]
gi|20978795|sp|Q9XEC3|WR42_ARATH Probable WRKY
transcription factor 42 (WRKY DNA-binding protein 42)
gi|25407272|pir||C85056 probable DNA-binding protein
[imported] - Arabidopsis thaliana
gi|4773884|gb|AAD29757.1|AF076243_4 putative DNA-binding
protein [Arabidopsis thaliana]
gi|7267202|emb|CAB77913.1| putative DNA-binding protein
[Arabidopsis thaliana] gi|15990594|gb|AAL11011.1| WRKY
transcription factor 42 [Arabidopsis thaliana]
Length = 528
Score = 89.4 bits (220), Expect = 4e-17
Identities = 59/140 (42%), Positives = 78/140 (55%), Gaps = 8/140 (5%)
Frame = -3
Query: 666 SAPFPTVTLDLTHNPNP--------LQFSRPQHSAPFQIPQNFMSGPASFAQAAPLYNQS 511
SAPFPT+TLDLT +PN +QFS Q S ++ Q+ + P QA QS
Sbjct: 405 SAPFPTITLDLTESPNGNNPTNNPLMQFS--QRSGLVELNQSVL--PHMMGQALYYNQQS 460
Query: 510 KFSGLQLSSQQEVGSSHQLASQQPPQQQPSLADTVSAATAAITADPNFTAVLAAAISSII 331
KFSGL + P Q + ++VSAATAAI ++PNF A LAAAI+SII
Sbjct: 461 KFSGLHM-----------------PSQPLNAGESVSAATAAIASNPNFAAALAAAITSII 503
Query: 330 GGSHANNNGSHNNNNNNNNN 271
GS+ NG++NN+N +N
Sbjct: 504 NGSNNQQNGNNNNSNVTTSN 523
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 631,672,897
Number of Sequences: 1393205
Number of extensions: 16286731
Number of successful extensions: 431201
Number of sequences better than 10.0: 3450
Number of HSP's better than 10.0 without gapping: 96155
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 271317
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29138478756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)