Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003348A_C01 KMC003348A_c01
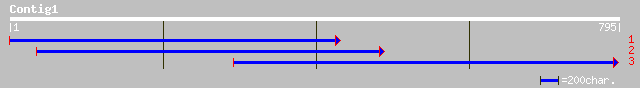
(795 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_177064.1| putative DNA-binding protein; protein id: At1g6... 247 2e-64
gb|AAM63847.1| putative DNA-binding protein [Arabidopsis thaliana] 246 2e-64
ref|NP_196430.1| myc-like protein; protein id: At5g08130.1 [Arab... 129 5e-29
dbj|BAB08642.1| DNA-binding protein-like [Arabidopsis thaliana] 128 8e-29
ref|NP_198702.1| putative protein; protein id: At5g38860.1 [Arab... 128 8e-29
>ref|NP_177064.1| putative DNA-binding protein; protein id: At1g69010.1, supported by
cDNA: 27793. [Arabidopsis thaliana]
gi|25404792|pir||E96714 probable DNA-binding protein
T6L1.19 [imported] - Arabidopsis thaliana
gi|12323222|gb|AAG51594.1|AC011665_15 putative
DNA-binding protein [Arabidopsis thaliana]
gi|26983866|gb|AAN86185.1| putative DNA-binding protein
[Arabidopsis thaliana]
Length = 311
Score = 247 bits (630), Expect = 2e-64
Identities = 121/184 (65%), Positives = 150/184 (80%), Gaps = 2/184 (1%)
Frame = +3
Query: 246 MKAGKSNQEEDEYEEEDFGSSQKHGPSS--APNTNKDAKATDKASAIRSKHSVTEQRRRS 419
M+ GK NQEE++Y EEDF +S++ GPSS ++N+D+K DKASAIRSKHSVTEQRRRS
Sbjct: 1 MRTGKGNQEEEDYGEEDF-NSKREGPSSNTTVHSNRDSKENDKASAIRSKHSVTEQRRRS 59
Query: 420 KINERFQILRDIIPHSEQKRDTASFLLEVIEYVQYLQEKVQKYEGSYPGWSQEPSKLMPW 599
KINERFQILR++IP+SEQKRDTASFLLEVI+YVQYLQEKVQKYEGSYPGWSQEP+KL PW
Sbjct: 60 KINERFQILRELIPNSEQKRDTASFLLEVIDYVQYLQEKVQKYEGSYPGWSQEPTKLTPW 119
Query: 600 RNSHWRVQNFVGQPPAVRNGSGPVSPFPAKFDDSNVSISPTMLSGTQNMIDPDQSRDILS 779
RN+HWRVQ+ P A+ NGSGP PFP KF+D+ V+ +P +++ Q I+ D++R I
Sbjct: 120 RNNHWRVQSLGNHPVAINNGSGPGIPFPGKFEDNTVTSTPAIIAEPQIPIESDKARAITG 179
Query: 780 KTAE 791
+ E
Sbjct: 180 ISIE 183
>gb|AAM63847.1| putative DNA-binding protein [Arabidopsis thaliana]
Length = 311
Score = 246 bits (629), Expect = 2e-64
Identities = 121/184 (65%), Positives = 150/184 (80%), Gaps = 2/184 (1%)
Frame = +3
Query: 246 MKAGKSNQEEDEYEEEDFGSSQKHGPSS--APNTNKDAKATDKASAIRSKHSVTEQRRRS 419
M+ GK NQEE++Y EEDF +S++ GPSS ++N+D+K DKASAIRSKHSVTEQRRRS
Sbjct: 1 MRTGKRNQEEEDYGEEDF-NSKREGPSSNTTVHSNRDSKENDKASAIRSKHSVTEQRRRS 59
Query: 420 KINERFQILRDIIPHSEQKRDTASFLLEVIEYVQYLQEKVQKYEGSYPGWSQEPSKLMPW 599
KINERFQILR++IP+SEQKRDTASFLLEVI+YVQYLQEKVQKYEGSYPGWSQEP+KL PW
Sbjct: 60 KINERFQILRELIPNSEQKRDTASFLLEVIDYVQYLQEKVQKYEGSYPGWSQEPTKLTPW 119
Query: 600 RNSHWRVQNFVGQPPAVRNGSGPVSPFPAKFDDSNVSISPTMLSGTQNMIDPDQSRDILS 779
RN+HWRVQ+ P A+ NGSGP PFP KF+D+ V+ +P +++ Q I+ D++R I
Sbjct: 120 RNNHWRVQSLGNHPVAINNGSGPGIPFPGKFEDNTVTSTPAIIAEPQIPIESDKARAITG 179
Query: 780 KTAE 791
+ E
Sbjct: 180 ISIE 183
>ref|NP_196430.1| myc-like protein; protein id: At5g08130.1 [Arabidopsis thaliana]
gi|11358566|pir||T50498 myc-like protein - Arabidopsis
thaliana gi|8346550|emb|CAB93714.1| myc-like protein
[Arabidopsis thaliana]
Length = 530
Score = 129 bits (324), Expect = 5e-29
Identities = 79/175 (45%), Positives = 105/175 (59%), Gaps = 8/175 (4%)
Frame = +3
Query: 111 PLSINSSTPFALNFSSGVSPFRSKFSSFEDSELVSNWGGTLLRATMKAGKSNQEEDEYEE 290
P++ S N SG S SS + S L S ++R+ A S+QE+D +E
Sbjct: 187 PVAERRSQSLTNNHMSGFS----SLSSSQGSVLKSQSFMDMIRS---AKGSSQEDDLDDE 239
Query: 291 EDF-------GSSQKHGPSSAPNTNKDAKATD-KASAIRSKHSVTEQRRRSKINERFQIL 446
EDF +SQ H + D K + RSKHS TEQRRRSKIN+RFQ+L
Sbjct: 240 EDFIMKKESSSTSQSHRVDLRVKADVRGSPNDQKLNTPRSKHSATEQRRRSKINDRFQML 299
Query: 447 RDIIPHSEQKRDTASFLLEVIEYVQYLQEKVQKYEGSYPGWSQEPSKLMPWRNSH 611
R +IP+S+QKRD ASFLLEVIEY+Q+LQEK KY SY GW+ EP+KL+ W++++
Sbjct: 300 RQLIPNSDQKRDKASFLLEVIEYIQFLQEKADKYVTSYQGWNHEPAKLLNWQSNN 354
>dbj|BAB08642.1| DNA-binding protein-like [Arabidopsis thaliana]
Length = 298
Score = 128 bits (322), Expect = 8e-29
Identities = 73/158 (46%), Positives = 93/158 (58%), Gaps = 2/158 (1%)
Frame = +3
Query: 324 SSAPNTNKDAKATDKASAIRSKHSVTEQRRRSKINERFQILRDIIPHSE--QKRDTASFL 497
S+ P+ N + ++ RSKHS TEQRRRSKINERFQ L DIIP ++ QKRD ASFL
Sbjct: 16 SNLPSRNDSSTGRRNRNSCRSKHSETEQRRRSKINERFQSLMDIIPQNQNDQKRDKASFL 75
Query: 498 LEVIEYVQYLQEKVQKYEGSYPGWSQEPSKLMPWRNSHWRVQNFVGQPPAVRNGSGPVSP 677
LEVIEY+ +LQEKV YE S+ W Q P+KL+PWRNSH V P V++ S
Sbjct: 76 LEVIEYIHFLQEKVHMYEDSHQMWYQSPTKLIPWRNSHGSVAEENDHPQIVKSFS----- 130
Query: 678 FPAKFDDSNVSISPTMLSGTQNMIDPDQSRDILSKTAE 791
+ V+ S L T N ++PD + +K E
Sbjct: 131 -----SNDKVAASSGFLLDTYNSVNPDIDSAVSTKIPE 163
>ref|NP_198702.1| putative protein; protein id: At5g38860.1 [Arabidopsis thaliana]
Length = 353
Score = 128 bits (322), Expect = 8e-29
Identities = 73/158 (46%), Positives = 93/158 (58%), Gaps = 2/158 (1%)
Frame = +3
Query: 324 SSAPNTNKDAKATDKASAIRSKHSVTEQRRRSKINERFQILRDIIPHSE--QKRDTASFL 497
S+ P+ N + ++ RSKHS TEQRRRSKINERFQ L DIIP ++ QKRD ASFL
Sbjct: 16 SNLPSRNDSSTGRRNRNSCRSKHSETEQRRRSKINERFQSLMDIIPQNQNDQKRDKASFL 75
Query: 498 LEVIEYVQYLQEKVQKYEGSYPGWSQEPSKLMPWRNSHWRVQNFVGQPPAVRNGSGPVSP 677
LEVIEY+ +LQEKV YE S+ W Q P+KL+PWRNSH V P V++ S
Sbjct: 76 LEVIEYIHFLQEKVHMYEDSHQMWYQSPTKLIPWRNSHGSVAEENDHPQIVKSFS----- 130
Query: 678 FPAKFDDSNVSISPTMLSGTQNMIDPDQSRDILSKTAE 791
+ V+ S L T N ++PD + +K E
Sbjct: 131 -----SNDKVAASSGFLLDTYNSVNPDIDSAVSTKIPE 163
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 697,237,064
Number of Sequences: 1393205
Number of extensions: 15605730
Number of successful extensions: 64240
Number of sequences better than 10.0: 771
Number of HSP's better than 10.0 without gapping: 57016
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 63213
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40055936206
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)