Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003339A_C01 KMC003339A_c01
(637 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
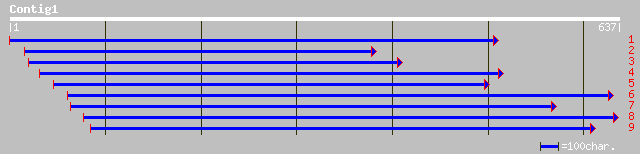
gb|AAK59482.2| unknown protein [Arabidopsis thaliana] 201 7e-51
ref|NP_565789.1| expressed protein; protein id: At2g34460.1, sup... 201 7e-51
pir||T02333 hypothetical protein At2g34460 [imported] - Arabidop... 201 7e-51
ref|NP_441422.1| hypothetical protein [Synechocystis sp. PCC 680... 107 1e-22
ref|NP_486791.1| hypothetical protein [Nostoc sp. PCC 7120] gi|2... 105 4e-22
>gb|AAK59482.2| unknown protein [Arabidopsis thaliana]
Length = 268
Score = 201 bits (511), Expect = 7e-51
Identities = 98/126 (77%), Positives = 116/126 (91%)
Frame = -1
Query: 634 INRFILVSSILVNGAAMGQLLNPAYIFLNVFGLTLVAKLQAENHIRKSGINYTIIRPGGL 455
+ +F+LVSSILVNGAAMGQ+LNPAY+FLN+FGLTLVAKLQAE +I+KSGINYTI+RPGGL
Sbjct: 141 VEKFVLVSSILVNGAAMGQILNPAYLFLNLFGLTLVAKLQAEKYIKKSGINYTIVRPGGL 200
Query: 454 RNDPPTGNVVMEPEDTLSQGSISRDHVAEVAVEALACPEASYKVVEIVSRPDAPKRTYHD 275
+NDPPTGNVVMEPEDTL +GSISRD VAEVAVEAL E+S+KVVEIV+R +APKR+Y D
Sbjct: 201 KNDPPTGNVVMEPEDTLYEGSISRDLVAEVAVEALLQEESSFKVVEIVARAEAPKRSYKD 260
Query: 274 LFGSIR 257
LF S++
Sbjct: 261 LFASVK 266
>ref|NP_565789.1| expressed protein; protein id: At2g34460.1, supported by cDNA:
gi_14334607, supported by cDNA: gi_15912294, supported
by cDNA: gi_17065635 [Arabidopsis thaliana]
gi|15912295|gb|AAL08281.1| At2g34460/T31E10.20
[Arabidopsis thaliana] gi|20197081|gb|AAC26697.2|
expressed protein [Arabidopsis thaliana]
gi|20197169|gb|AAM14955.1| expressed protein
[Arabidopsis thaliana]
Length = 246
Score = 201 bits (511), Expect = 7e-51
Identities = 98/126 (77%), Positives = 116/126 (91%)
Frame = -1
Query: 634 INRFILVSSILVNGAAMGQLLNPAYIFLNVFGLTLVAKLQAENHIRKSGINYTIIRPGGL 455
+ +F+LVSSILVNGAAMGQ+LNPAY+FLN+FGLTLVAKLQAE +I+KSGINYTI+RPGGL
Sbjct: 119 VEKFVLVSSILVNGAAMGQILNPAYLFLNLFGLTLVAKLQAEKYIKKSGINYTIVRPGGL 178
Query: 454 RNDPPTGNVVMEPEDTLSQGSISRDHVAEVAVEALACPEASYKVVEIVSRPDAPKRTYHD 275
+NDPPTGNVVMEPEDTL +GSISRD VAEVAVEAL E+S+KVVEIV+R +APKR+Y D
Sbjct: 179 KNDPPTGNVVMEPEDTLYEGSISRDLVAEVAVEALLQEESSFKVVEIVARAEAPKRSYKD 238
Query: 274 LFGSIR 257
LF S++
Sbjct: 239 LFASVK 244
>pir||T02333 hypothetical protein At2g34460 [imported] - Arabidopsis thaliana
gi|24030209|gb|AAN41284.1| unknown protein [Arabidopsis
thaliana]
Length = 280
Score = 201 bits (511), Expect = 7e-51
Identities = 98/126 (77%), Positives = 116/126 (91%)
Frame = -1
Query: 634 INRFILVSSILVNGAAMGQLLNPAYIFLNVFGLTLVAKLQAENHIRKSGINYTIIRPGGL 455
+ +F+LVSSILVNGAAMGQ+LNPAY+FLN+FGLTLVAKLQAE +I+KSGINYTI+RPGGL
Sbjct: 153 VEKFVLVSSILVNGAAMGQILNPAYLFLNLFGLTLVAKLQAEKYIKKSGINYTIVRPGGL 212
Query: 454 RNDPPTGNVVMEPEDTLSQGSISRDHVAEVAVEALACPEASYKVVEIVSRPDAPKRTYHD 275
+NDPPTGNVVMEPEDTL +GSISRD VAEVAVEAL E+S+KVVEIV+R +APKR+Y D
Sbjct: 213 KNDPPTGNVVMEPEDTLYEGSISRDLVAEVAVEALLQEESSFKVVEIVARAEAPKRSYKD 272
Query: 274 LFGSIR 257
LF S++
Sbjct: 273 LFASVK 278
>ref|NP_441422.1| hypothetical protein [Synechocystis sp. PCC 6803]
gi|7450531|pir||S75541 hypothetical protein sll1218 -
Synechocystis sp. (strain PCC 6803)
gi|1653186|dbj|BAA18102.1| ORF_ID:sll1218~hypothetical
protein [Synechocystis sp. PCC 6803]
Length = 219
Score = 107 bits (268), Expect = 1e-22
Identities = 59/126 (46%), Positives = 81/126 (63%)
Frame = -1
Query: 634 INRFILVSSILVNGAAMGQLLNPAYIFLNVFGLTLVAKLQAENHIRKSGINYTIIRPGGL 455
I + +LVSS+ V+ L +P LN+FGL LV K EN++R+SG+ YTI+RPGGL
Sbjct: 103 IEQLVLVSSLCVSN-----LFHP----LNLFGLILVWKQWGENYLRQSGVPYTIVRPGGL 153
Query: 454 RNDPPTGNVVMEPEDTLSQGSISRDHVAEVAVEALACPEASYKVVEIVSRPDAPKRTYHD 275
+N+ +VM DTL GSI R VAE VE+L P A K+VEIVS+PD P +++ +
Sbjct: 154 KNEDNDNAIVMAGADTLFDGSIPRQKVAEACVESLFSPSAKNKIVEIVSKPDIPVQSFDE 213
Query: 274 LFGSIR 257
LF +R
Sbjct: 214 LFAMVR 219
>ref|NP_486791.1| hypothetical protein [Nostoc sp. PCC 7120] gi|25327951|pir||AH2149
hypothetical protein alr2751 [imported] - Nostoc sp.
(strain PCC 7120) gi|17131844|dbj|BAB74450.1|
ORF_ID:alr2751~hypothetical protein [Nostoc sp. PCC
7120]
Length = 218
Score = 105 bits (263), Expect = 4e-22
Identities = 57/122 (46%), Positives = 83/122 (67%)
Frame = -1
Query: 634 INRFILVSSILVNGAAMGQLLNPAYIFLNVFGLTLVAKLQAENHIRKSGINYTIIRPGGL 455
I F+LV+S+ V+ Q +P LN+F L LV K QAE +++KSG+ YTI+RPGGL
Sbjct: 103 IENFVLVTSLCVS-----QFFHP----LNLFWLILVWKKQAEEYLQKSGLTYTIVRPGGL 153
Query: 454 RNDPPTGNVVMEPEDTLSQGSISRDHVAEVAVEALACPEASYKVVEIVSRPDAPKRTYHD 275
+N+ + +VM+ DTL GSI R VA+V VE+L P+A K+VEIV++P+A +T+ +
Sbjct: 154 KNEDNSDAIVMQSSDTLFDGSIPRQKVAQVCVESLFEPDARNKIVEIVAKPEASSKTFTE 213
Query: 274 LF 269
LF
Sbjct: 214 LF 215
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 514,678,932
Number of Sequences: 1393205
Number of extensions: 10860537
Number of successful extensions: 27547
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 26578
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27503
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)