Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
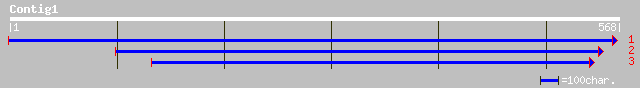
Query= KMC003332A_C01 KMC003332A_c01
(568 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_623872.1| Diguanylate cyclase/phosphodiesterase domain 1 ... 34 1.4
ref|NP_690500.1| cytochrome-c oxidase subunit III [Heliothis zea... 33 3.2
ref|NP_732478.1| CG31210-PA [Drosophila melanogaster] gi|2317176... 32 7.2
gb|AAL74352.1| s48/45 [Plasmodium falciparum] 31 9.4
gb|AAL74350.1| s48/45 [Plasmodium falciparum] 31 9.4
>ref|NP_623872.1| Diguanylate cyclase/phosphodiesterase domain 1 (GGDEF)
[Thermoanaerobacter tengcongensis]
gi|20517340|gb|AAM25476.1| Diguanylate
cyclase/phosphodiesterase domain 1 (GGDEF)
[Thermoanaerobacter tengcongensis]
Length = 553
Score = 33.9 bits (76), Expect = 1.4
Identities = 28/81 (34%), Positives = 41/81 (50%), Gaps = 2/81 (2%)
Frame = -1
Query: 469 SIKETKTILVRMFCSALIHGNTFT*SLEMEGWSVFLEIWTHS--LSLS*FSLSLP*SHML 296
SI + K ILV + S LI+ +L+++G +F E W S L L + L +P + +L
Sbjct: 129 SIAQVKYILVYVIVSFLINNFILYYALKVQGKILFKEYWNESVLLELGTYVLMVPAAMLL 188
Query: 295 A*LIFYIVASSSLQLEVLSST 233
A Y+ SL VLS T
Sbjct: 189 A----YVYFKHSLMFFVLSLT 205
>ref|NP_690500.1| cytochrome-c oxidase subunit III [Heliothis zea virus 1]
gi|22671548|gb|AAN04375.1|AF451898_80 cytochrome-c
oxidase subunit III [Heliothis zea virus 1]
Length = 126
Score = 32.7 bits (73), Expect = 3.2
Identities = 26/74 (35%), Positives = 35/74 (47%), Gaps = 1/74 (1%)
Frame = -2
Query: 270 PLH-LYNLKSFQVLGSLWRIDYSMDTFFIYLMFLFFNLHVHIETFGILLDLK*GVRYILP 94
PLH + LK L S ID S + F + F+N +VH + F I+ GV YIL
Sbjct: 46 PLHAMLKLKQNDALNSTCAIDLSFECFCV---LCFYNFYVHFQGFCIVF----GVSYILC 98
Query: 93 LRLI*KYIF*KPYT 52
L ++F YT
Sbjct: 99 FLL--DFVFFSNYT 110
>ref|NP_732478.1| CG31210-PA [Drosophila melanogaster] gi|23171766|gb|AAN13818.1|
CG31210-PA [Drosophila melanogaster]
Length = 130
Score = 31.6 bits (70), Expect = 7.2
Identities = 14/45 (31%), Positives = 22/45 (48%)
Frame = -2
Query: 444 WCVCFVQLSYMAIPLRDHLRWRGGQFFLKFGHIPLAFLSFLSHFH 310
W C + L++ ++P H+ + G Q + GHI A L H H
Sbjct: 66 WSYCILVLAFQSLPQIGHIHFEG-QLHRRNGHIVEAIAQILRHIH 109
>gb|AAL74352.1| s48/45 [Plasmodium falciparum]
Length = 448
Score = 31.2 bits (69), Expect = 9.4
Identities = 15/44 (34%), Positives = 23/44 (52%)
Frame = +2
Query: 92 NGSIYLTPYFKSSNMPKVSICTCKLKNKNIK*IKKVSME*SILH 223
N +I+ P++ +S CTCK KN N K + K E ++H
Sbjct: 254 NLTIFKAPFYVTSKDVNTE-CTCKFKNNNYKIVLKPKYEKKVIH 296
>gb|AAL74350.1| s48/45 [Plasmodium falciparum]
Length = 448
Score = 31.2 bits (69), Expect = 9.4
Identities = 15/44 (34%), Positives = 23/44 (52%)
Frame = +2
Query: 92 NGSIYLTPYFKSSNMPKVSICTCKLKNKNIK*IKKVSME*SILH 223
N +I+ P++ +S CTCK KN N K + K E ++H
Sbjct: 254 NLTIFKAPFYVTSKDVNTE-CTCKFKNNNYKIVLKPKYEKKVIH 296
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 462,109,858
Number of Sequences: 1393205
Number of extensions: 9366737
Number of successful extensions: 25665
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 23267
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25532
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)