Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003322A_C02 KMC003322A_c02
(910 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
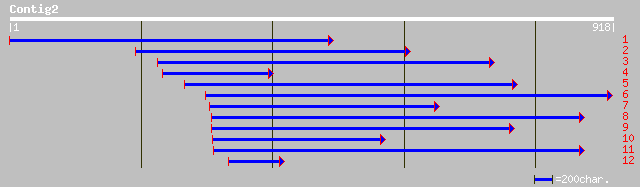
emb|CAA98160.1| RAB1C [Lotus japonicus] 244 1e-63
dbj|BAA76422.1| rab-type small GTP-binding protein [Cicer arieti... 237 2e-61
dbj|BAA02116.1| GTP-binding protein [Pisum sativum] gi|738940|pr... 231 1e-59
gb|AAA80678.1| small GTP-binding protein 228 1e-58
emb|CAA69701.1| small GTP-binding protein [Nicotiana plumbaginif... 224 2e-57
>emb|CAA98160.1| RAB1C [Lotus japonicus]
Length = 202
Score = 244 bits (624), Expect = 1e-63
Identities = 121/122 (99%), Positives = 121/122 (99%)
Frame = -3
Query: 908 AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKCDLTANKVVSYETAKAFA 729
AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKCDLT NKVVSYETAKAFA
Sbjct: 81 AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKCDLTENKVVSYETAKAFA 140
Query: 728 DEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQPVNNARPPTVQIRGQPVNQKSGCC 549
DEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQPVNNARPPTVQIRGQPVNQKSGCC
Sbjct: 141 DEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQPVNNARPPTVQIRGQPVNQKSGCC 200
Query: 548 SS 543
SS
Sbjct: 201 SS 202
>dbj|BAA76422.1| rab-type small GTP-binding protein [Cicer arietinum]
Length = 202
Score = 237 bits (605), Expect = 2e-61
Identities = 117/122 (95%), Positives = 119/122 (96%)
Frame = -3
Query: 908 AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKCDLTANKVVSYETAKAFA 729
AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKCDL ANKVVS ETAKAFA
Sbjct: 81 AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKCDLAANKVVSSETAKAFA 140
Query: 728 DEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQPVNNARPPTVQIRGQPVNQKSGCC 549
DEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQP NNARPPTVQIRGQP+NQKSGCC
Sbjct: 141 DEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQPANNARPPTVQIRGQPMNQKSGCC 200
Query: 548 SS 543
S+
Sbjct: 201 ST 202
>dbj|BAA02116.1| GTP-binding protein [Pisum sativum] gi|738940|prf||2001457H
GTP-binding protein
Length = 202
Score = 231 bits (588), Expect = 1e-59
Identities = 116/122 (95%), Positives = 116/122 (95%)
Frame = -3
Query: 908 AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKCDLTANKVVSYETAKAFA 729
AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNK DL NKVVS ETAKAFA
Sbjct: 81 AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKSDLADNKVVSSETAKAFA 140
Query: 728 DEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQPVNNARPPTVQIRGQPVNQKSGCC 549
DEIGIPFMETSAKNA NVEQAFMAMAAEIKNRMASQP NNARPPTVQIRGQPVNQKSGCC
Sbjct: 141 DEIGIPFMETSAKNANNVEQAFMAMAAEIKNRMASQPSNNARPPTVQIRGQPVNQKSGCC 200
Query: 548 SS 543
SS
Sbjct: 201 SS 202
>gb|AAA80678.1| small GTP-binding protein
Length = 203
Score = 228 bits (581), Expect = 1e-58
Identities = 115/123 (93%), Positives = 118/123 (95%), Gaps = 1/123 (0%)
Frame = -3
Query: 908 AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKCDLTANKVVSYETAKAFA 729
AHGIIVVYDVTDQESFNNVKQWL+EIDRYAS+NVNKLLVGNKCDLTA KVVS ETA+AFA
Sbjct: 81 AHGIIVVYDVTDQESFNNVKQWLSEIDRYASDNVNKLLVGNKCDLTAQKVVSTETAQAFA 140
Query: 728 DEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQPV-NNARPPTVQIRGQPVNQKSGC 552
DEIGIPFMETSAKNATNVEQAFMAMAA IKNRMASQP NNARPPTVQIRGQPVNQKSGC
Sbjct: 141 DEIGIPFMETSAKNATNVEQAFMAMAASIKNRMASQPASNNARPPTVQIRGQPVNQKSGC 200
Query: 551 CSS 543
CSS
Sbjct: 201 CSS 203
>emb|CAA69701.1| small GTP-binding protein [Nicotiana plumbaginifolia]
Length = 203
Score = 224 bits (570), Expect = 2e-57
Identities = 113/123 (91%), Positives = 117/123 (94%), Gaps = 1/123 (0%)
Frame = -3
Query: 908 AHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKCDLTANKVVSYETAKAFA 729
AHGIIVVYDVTDQESFNNVKQWL+EIDRYAS++VNKLLVGNKCDLTA KVVS ETA+AFA
Sbjct: 81 AHGIIVVYDVTDQESFNNVKQWLSEIDRYASDSVNKLLVGNKCDLTAQKVVSTETAQAFA 140
Query: 728 DEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQPV-NNARPPTVQIRGQPVNQKSGC 552
DEIGIPFMETSAKNATNVEQAFMAMAA IKNRMASQP NNARPPTVQIRGQPVNQKSGC
Sbjct: 141 DEIGIPFMETSAKNATNVEQAFMAMAASIKNRMASQPASNNARPPTVQIRGQPVNQKSGC 200
Query: 551 CSS 543
C S
Sbjct: 201 CLS 203
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 757,476,161
Number of Sequences: 1393205
Number of extensions: 16237906
Number of successful extensions: 55055
Number of sequences better than 10.0: 1447
Number of HSP's better than 10.0 without gapping: 52115
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 54513
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 49641180728
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)