Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
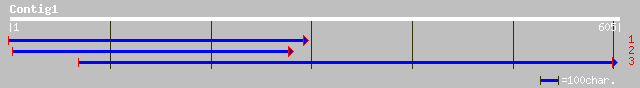
Query= KMC003313A_C01 KMC003313A_c01
(605 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_014280.1| Hypothetical ORF; Ynl119wp [Saccharomyces cerev... 35 0.58
ref|NP_764278.1| conserved hypothetical protein [Staphylococcus ... 35 0.76
gb|EAA15614.1| Unknown protein [Plasmodium yoelii yoelii] 32 6.4
gb|AAF25576.1|AF120104_1 mucus binding protein precursor; Mub [L... 32 6.4
ref|NP_704630.1| hypothetical protein [Plasmodium falciparum 3D7... 32 8.4
>ref|NP_014280.1| Hypothetical ORF; Ynl119wp [Saccharomyces cerevisiae]
gi|1730768|sp|P53923|YNL9_YEAST Hypothetical 56.5 kDa
protein in TOM70-PSU1 intergenic region
gi|2131893|pir||S63060 hypothetical protein YNL119w -
yeast (Saccharomyces cerevisiae)
gi|1183954|emb|CAA93388.1| N1913 [Saccharomyces
cerevisiae] gi|1302048|emb|CAA96001.1| ORF YNL119w
[Saccharomyces cerevisiae]
Length = 493
Score = 35.4 bits (80), Expect = 0.58
Identities = 26/79 (32%), Positives = 38/79 (47%), Gaps = 6/79 (7%)
Frame = +1
Query: 34 NFLATIFRVHQIILNSLCTAASSIYTMTTENGTFNML*K---ITTS*TQTMKGIQDR--- 195
N ++ IF VH I +N AS + T +N F + K + S T+K I +
Sbjct: 132 NKISNIFNVHFIDVNEFFNNASEVSTFIIDNENFEIFSKSKSVDDSNILTLKEILGKYCL 191
Query: 196 NRTTLRQCHSFIKTQLIRH 252
N ++ S IKTQLI+H
Sbjct: 192 NNSSRSDLISIIKTQLIKH 210
>ref|NP_764278.1| conserved hypothetical protein [Staphylococcus epidermidis ATCC
12228] gi|27315185|gb|AAO04320.1|AE016746_110 conserved
hypothetical protein [Staphylococcus epidermidis ATCC
12228]
Length = 268
Score = 35.0 bits (79), Expect = 0.76
Identities = 13/37 (35%), Positives = 26/37 (70%)
Frame = -2
Query: 511 LNQHEEMSEEGNKKGKQNLPPPQVQLRDIGDWDDSQR 401
+N ++++E KGK N P +++++++GD DDS+R
Sbjct: 151 INDVRKIAKELEHKGKVNYPNTEIKIKNVGDLDDSER 187
>gb|EAA15614.1| Unknown protein [Plasmodium yoelii yoelii]
Length = 635
Score = 32.0 bits (71), Expect = 6.4
Identities = 14/40 (35%), Positives = 21/40 (52%)
Frame = -2
Query: 568 RLFRNIGDQPSSSAGKSGHLNQHEEMSEEGNKKGKQNLPP 449
+L++NI Q KS +N + +SEEGNK + P
Sbjct: 169 KLYKNIESQKKIIKNKSNEINSIQHLSEEGNKNDNMDKDP 208
>gb|AAF25576.1|AF120104_1 mucus binding protein precursor; Mub [Lactobacillus reuteri]
Length = 3269
Score = 32.0 bits (71), Expect = 6.4
Identities = 20/63 (31%), Positives = 28/63 (43%), Gaps = 1/63 (1%)
Frame = -2
Query: 553 IGDQPSSSAGKSGHLNQHEEMSEEGNKKGKQNLPPPQVQLRDIGDWD-DSQRPPKFQQHL 377
+ DQ GK+G H E ++ K GKQ Q D +D D+Q P +F +L
Sbjct: 572 LNDQTIQLTGKTGEKISHTEANQTLAKLGKQGYVVDQNTFADDATYDNDTQAPQEFTIYL 631
Query: 376 NED 368
D
Sbjct: 632 KHD 634
>ref|NP_704630.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23504990|emb|CAD51773.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 1012
Score = 31.6 bits (70), Expect = 8.4
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 5/50 (10%)
Frame = +3
Query: 84 LYCCIIHLYNDN*KWHL*HALENNYKLNTN-----NERNPRS*QNNIKTM 218
LY CI++++ DN LENN K+ TN E N + QNNI +
Sbjct: 106 LYKCILNIFEDN------EILENNNKIETNINKQIIEHNKNNIQNNINNI 149
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 496,987,451
Number of Sequences: 1393205
Number of extensions: 10404415
Number of successful extensions: 28746
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 26115
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28396
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)