Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003306A_C01 KMC003306A_c01
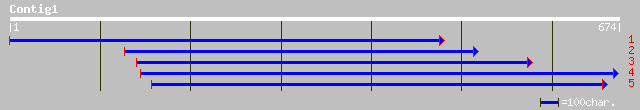
(674 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_187434.1| putative heat-shock protein; protein id: At3g07... 162 4e-39
pir||S49340 heat-shock protein, 82K, precursor - rye gi|556673|e... 159 3e-38
ref|NP_178487.1| putative heat shock protein; protein id: At2g04... 148 6e-35
gb|AAM19795.1| At2g04030/F3C11.14 [Arabidopsis thaliana] 148 6e-35
emb|CAA72515.1| heat shock protein [Arabidopsis thaliana] 145 5e-34
>ref|NP_187434.1| putative heat-shock protein; protein id: At3g07770.1 [Arabidopsis
thaliana] gi|6466963|gb|AAF13098.1|AC009176_25 putative
heat-shock protein [Arabidopsis thaliana]
gi|6648189|gb|AAF21187.1|AC013483_11 putative heat-shock
protein [Arabidopsis thaliana]
Length = 803
Score = 162 bits (410), Expect = 4e-39
Identities = 81/127 (63%), Positives = 99/127 (77%), Gaps = 2/127 (1%)
Frame = -2
Query: 673 NMERLMKPQSMGDASSMDFMRSRRVFEINPDHPIIRNLDAACKTNPDDQEAERAIDLLYD 494
NMERLMK QS GD S+D+M+ RRVFEINPDH II+N++AA +NP+D++A RAIDL+YD
Sbjct: 677 NMERLMKAQSTGDTISLDYMKGRRVFEINPDHSIIKNINAAYNSNPNDEDAMRAIDLMYD 736
Query: 493 AALVSSGYTPDNPAQLGGKIYEMMSMALSGRWSAS--GQFQSTVTQPPVPEFVEAEVVEP 320
AALVSSG+TPDNPA+LGGKIYEMM +ALSG+WS+ Q + E EAEVVEP
Sbjct: 737 AALVSSGFTPDNPAELGGKIYEMMDVALSGKWSSPEVQPQQQQMAHSHDAETFEAEVVEP 796
Query: 319 TEADGQK 299
E DG+K
Sbjct: 797 VEVDGKK 803
>pir||S49340 heat-shock protein, 82K, precursor - rye gi|556673|emb|CAA82945.1|
heat-shock protein [Secale cereale]
Length = 781
Score = 159 bits (403), Expect = 3e-38
Identities = 77/133 (57%), Positives = 100/133 (74%), Gaps = 1/133 (0%)
Frame = -2
Query: 673 NMERLMKPQSMGDASSMDFMRSRRVFEINPDHPIIRNLDAACKTNPDDQEAERAIDLLYD 494
NMERLMK Q++GD SS++FMR RR+FEINPDHPI+++L AACK PD EA+RA++LLY+
Sbjct: 649 NMERLMKAQTLGDTSSLEFMRGRRIFEINPDHPIVKDLSAACKNEPDSTEAKRAVELLYE 708
Query: 493 AALVSSGYTPDNPAQLGGKIYEMMSMALSGRWSASGQFQS-TVTQPPVPEFVEAEVVEPT 317
AL+SSGYTP++PA+LGGKIYEMM++AL GRW SG ++ T E V EV+EP+
Sbjct: 709 TALISSGYTPESPAELGGKIYEMMTIALGGRWGRSGAEEAETNVDGDSSEGVVPEVIEPS 768
Query: 316 EADGQK*EDCQRD 278
E + D RD
Sbjct: 769 EVRTENENDPWRD 781
>ref|NP_178487.1| putative heat shock protein; protein id: At2g04030.1, supported by
cDNA: gi_15450722, supported by cDNA: gi_16930684,
supported by cDNA: gi_20453105 [Arabidopsis thaliana]
gi|25295988|pir||H84453 probable heat shock protein
[imported] - Arabidopsis thaliana
gi|4914387|gb|AAD32922.1| putative heat shock protein
[Arabidopsis thaliana] gi|15450723|gb|AAK96633.1|
At2g04030/F3C11.14 [Arabidopsis thaliana]
gi|16930685|gb|AAL32008.1|AF436826_1 At2g04030/F3C11.14
[Arabidopsis thaliana] gi|25090168|gb|AAN72245.1|
At2g04030/F3C11.14 [Arabidopsis thaliana]
Length = 780
Score = 148 bits (374), Expect = 6e-35
Identities = 70/121 (57%), Positives = 92/121 (75%), Gaps = 1/121 (0%)
Frame = -2
Query: 673 NMERLMKPQSMGDASSMDFMRSRRVFEINPDHPIIRNLDAACKTNPDDQEAERAIDLLYD 494
NMERLMK Q++GD SS++FMR RR+ EINPDHPII++L+AACK P+ EA R +DLLYD
Sbjct: 650 NMERLMKAQALGDTSSLEFMRGRRILEINPDHPIIKDLNAACKNAPESTEATRVVDLLYD 709
Query: 493 AALVSSGYTPDNPAQLGGKIYEMMSMALSGRWS-ASGQFQSTVTQPPVPEFVEAEVVEPT 317
A++SSG+TPD+PA+LG KIYEMM+MA+ GRW + +S+ + E EVVEP+
Sbjct: 710 TAIISSGFTPDSPAELGNKIYEMMAMAVGGRWGRVEEEEESSTVNEGDDKSGETEVVEPS 769
Query: 316 E 314
E
Sbjct: 770 E 770
>gb|AAM19795.1| At2g04030/F3C11.14 [Arabidopsis thaliana]
Length = 780
Score = 148 bits (374), Expect = 6e-35
Identities = 70/121 (57%), Positives = 92/121 (75%), Gaps = 1/121 (0%)
Frame = -2
Query: 673 NMERLMKPQSMGDASSMDFMRSRRVFEINPDHPIIRNLDAACKTNPDDQEAERAIDLLYD 494
NMERLMK Q++GD SS++FMR RR+ EINPDHPII++L+AACK P+ EA R +DLLYD
Sbjct: 650 NMERLMKAQALGDTSSLEFMRGRRILEINPDHPIIKDLNAACKNAPESTEATRVVDLLYD 709
Query: 493 AALVSSGYTPDNPAQLGGKIYEMMSMALSGRWS-ASGQFQSTVTQPPVPEFVEAEVVEPT 317
A++SSG+TPD+PA+LG KIYEMM+MA+ GRW + +S+ + E EVVEP+
Sbjct: 710 TAIISSGFTPDSPAELGNKIYEMMAMAVGGRWGRVEEEEESSTVNEGDDKSGETEVVEPS 769
Query: 316 E 314
E
Sbjct: 770 E 770
>emb|CAA72515.1| heat shock protein [Arabidopsis thaliana]
Length = 768
Score = 145 bits (366), Expect = 5e-34
Identities = 68/121 (56%), Positives = 91/121 (75%), Gaps = 1/121 (0%)
Frame = -2
Query: 673 NMERLMKPQSMGDASSMDFMRSRRVFEINPDHPIIRNLDAACKTNPDDQEAERAIDLLYD 494
NMERLMK Q++GD SS++FMR RR+ EINPDHPII++L+AACK P+ EA R +DLLYD
Sbjct: 638 NMERLMKAQALGDTSSLEFMRGRRILEINPDHPIIKDLNAACKNAPESTEATRVVDLLYD 697
Query: 493 AALVSSGYTPDNPAQLGGKIYEMMSMALSGRWS-ASGQFQSTVTQPPVPEFVEAEVVEPT 317
A++S G+TPD+PA+LG KIYEMM++A+ GRW + +S+ + E EVVEP+
Sbjct: 698 TAIISGGFTPDSPAELGNKIYEMMAVAVGGRWGRVEEEEESSTVNEGDDKSGETEVVEPS 757
Query: 316 E 314
E
Sbjct: 758 E 758
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 589,509,260
Number of Sequences: 1393205
Number of extensions: 12336897
Number of successful extensions: 36065
Number of sequences better than 10.0: 283
Number of HSP's better than 10.0 without gapping: 34697
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36029
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29704274460
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)