Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
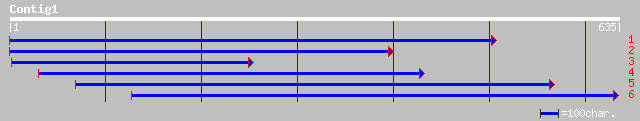
Query= KMC003292A_C01 KMC003292A_c01
(628 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_198955.1| putative protein; protein id: At5g41390.1 [Arab... 77 1e-13
pir||T05385 hypothetical protein F16G20.170 - Arabidopsis thalia... 77 2e-13
ref|NP_176568.1| unknown protein; protein id: At1g63830.1, suppo... 76 4e-13
ref|NP_194078.2| expressed protein; protein id: At4g23470.1, sup... 75 5e-13
gb|AAM63607.1| unknown [Arabidopsis thaliana] 75 5e-13
>ref|NP_198955.1| putative protein; protein id: At5g41390.1 [Arabidopsis thaliana]
gi|9758048|dbj|BAB08511.1|
gb|AAF07369.1~gene_id:MYC6.10~strong similarity to
unknown protein [Arabidopsis thaliana]
Length = 297
Score = 77.4 bits (189), Expect = 1e-13
Identities = 34/44 (77%), Positives = 36/44 (81%), Gaps = 1/44 (2%)
Frame = -3
Query: 473 CASYLLPKRALYN-MSRYTCCAGYMPCSKRCGESKCLEFCFCTE 345
C SYLL KRALYN MSRYTCC GYMPCS +CGESKC +FC TE
Sbjct: 45 CVSYLLRKRALYNDMSRYTCCGGYMPCSGKCGESKCPQFCLATE 88
>pir||T05385 hypothetical protein F16G20.170 - Arabidopsis thaliana
gi|3451072|emb|CAA20468.1| putative protein [Arabidopsis
thaliana] gi|7269195|emb|CAB79302.1| putative protein
[Arabidopsis thaliana]
Length = 160
Score = 77.0 bits (188), Expect = 2e-13
Identities = 35/47 (74%), Positives = 38/47 (80%), Gaps = 1/47 (2%)
Frame = -3
Query: 482 SAQCASYLLPKRALYN-MSRYTCCAGYMPCSKRCGESKCLEFCFCTE 345
SA CASYLL KRALY+ MSRY CCAGYMPCS RCGE+KC + C TE
Sbjct: 40 SAPCASYLLRKRALYDDMSRYVCCAGYMPCSGRCGEAKCPQLCLATE 86
>ref|NP_176568.1| unknown protein; protein id: At1g63830.1, supported by cDNA:
gi_19424092 [Arabidopsis thaliana]
gi|25404446|pir||D96663 unknown protein, 55304-53614
[imported] - Arabidopsis thaliana
gi|12325014|gb|AAG52456.1|AC010852_13 unknown protein;
55304-53614 [Arabidopsis thaliana]
gi|19424093|gb|AAL87329.1| unknown protein [Arabidopsis
thaliana] gi|21436183|gb|AAM51379.1| unknown protein
[Arabidopsis thaliana]
Length = 232
Score = 75.9 bits (185), Expect = 4e-13
Identities = 33/44 (75%), Positives = 36/44 (81%), Gaps = 1/44 (2%)
Frame = -3
Query: 473 CASYLLPKRALYN-MSRYTCCAGYMPCSKRCGESKCLEFCFCTE 345
C SY+L +RALYN MSRYTCCAGYMPCS RCGESKC + C TE
Sbjct: 45 CVSYMLRRRALYNDMSRYTCCAGYMPCSGRCGESKCPQLCLATE 88
>ref|NP_194078.2| expressed protein; protein id: At4g23470.1, supported by cDNA:
25694., supported by cDNA: gi_17065515, supported by
cDNA: gi_20148522 [Arabidopsis thaliana]
gi|17065516|gb|AAL32912.1| Unknown protein [Arabidopsis
thaliana] gi|20148523|gb|AAM10152.1| unknown protein
[Arabidopsis thaliana]
Length = 255
Score = 75.5 bits (184), Expect = 5e-13
Identities = 34/46 (73%), Positives = 37/46 (79%), Gaps = 1/46 (2%)
Frame = -3
Query: 479 AQCASYLLPKRALYN-MSRYTCCAGYMPCSKRCGESKCLEFCFCTE 345
A CASYLL KRALY+ MSRY CCAGYMPCS RCGE+KC + C TE
Sbjct: 41 APCASYLLRKRALYDDMSRYVCCAGYMPCSGRCGEAKCPQLCLATE 86
>gb|AAM63607.1| unknown [Arabidopsis thaliana]
Length = 247
Score = 75.5 bits (184), Expect = 5e-13
Identities = 34/46 (73%), Positives = 37/46 (79%), Gaps = 1/46 (2%)
Frame = -3
Query: 479 AQCASYLLPKRALYN-MSRYTCCAGYMPCSKRCGESKCLEFCFCTE 345
A CASYLL KRALY+ MSRY CCAGYMPCS RCGE+KC + C TE
Sbjct: 33 APCASYLLRKRALYDDMSRYVCCAGYMPCSGRCGEAKCPQLCLATE 78
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 522,121,775
Number of Sequences: 1393205
Number of extensions: 11084826
Number of successful extensions: 27193
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 25957
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27137
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25586195130
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)