Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003275A_C01 KMC003275A_c01
(549 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
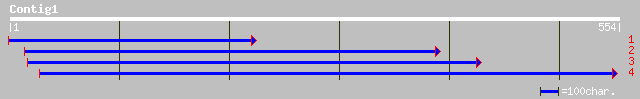
ref|NP_175875.1| hypothetical protein; protein id: At1g54770.1 [... 102 4e-21
ref|NP_594087.1| hypothetical protein [Schizosaccharomyces pombe... 78 6e-14
emb|CAB92796.1| dJ561L24.2 (acidic 82 kDa protein) [Homo sapiens] 78 6e-14
ref|NP_722501.1| hypothetical protein MGC27716 [Mus musculus] gi... 78 8e-14
ref|XP_215688.1| similar to hypothetical protein MGC27716 [Mus m... 77 1e-13
>ref|NP_175875.1| hypothetical protein; protein id: At1g54770.1 [Arabidopsis
thaliana] gi|27754411|gb|AAO22654.1| unknown protein
[Arabidopsis thaliana] gi|28393935|gb|AAO42375.1|
unknown protein [Arabidopsis thaliana]
Length = 189
Score = 102 bits (253), Expect = 4e-21
Identities = 52/103 (50%), Positives = 68/103 (65%)
Frame = -2
Query: 476 LILKQLRGAIDPKRHYKKGDSKSKTLPKYFQMGTVVDSPLDFFSGRLTKKERRASLADEL 297
L L +LR +DP HYKK S+SK KYFQ+GTV++ P + F GRLTKK R+A+LADEL
Sbjct: 87 LQLLKLRTVMDPALHYKKSVSRSKLAEKYFQIGTVIE-PAEEFYGRLTKKNRKATLADEL 145
Query: 296 LADQNLVAYRKRKVREIEEQNKPAGNENWKIKGGSCPEASKRK 168
++D YRKRKVREIEE+++ N+ W KG R+
Sbjct: 146 VSDPKTALYRKRKVREIEEKSRAVTNKKWNKKGNQSKNTKPRR 188
>ref|NP_594087.1| hypothetical protein [Schizosaccharomyces pombe]
gi|7491110|pir||T11651 hypothetical protein SPAC3G9.15c
- fission yeast (Schizosaccharomyces pombe)
gi|2706466|emb|CAA15924.1| eukarotic conserved protein;
similar to S. cerevisiae YLR051C [Schizosaccharomyces
pombe]
Length = 230
Score = 78.2 bits (191), Expect = 6e-14
Identities = 35/88 (39%), Positives = 59/88 (66%)
Frame = -2
Query: 470 LKQLRGAIDPKRHYKKGDSKSKTLPKYFQMGTVVDSPLDFFSGRLTKKERRASLADELLA 291
L ++R A+DPKRHY++ ++KS +PKYFQ+G++V+ P DF+S R+ +ER+ ++ DELL
Sbjct: 133 LLKMRNALDPKRHYRRENTKS--MPKYFQVGSIVEGPQDFYSSRIPTRERKETIVDELLH 190
Query: 290 DQNLVAYRKRKVREIEEQNKPAGNENWK 207
D +Y K+K E+++ +K
Sbjct: 191 DSERRSYFKKKYLELQKSKMSGRKGQYK 218
>emb|CAB92796.1| dJ561L24.2 (acidic 82 kDa protein) [Homo sapiens]
Length = 756
Score = 78.2 bits (191), Expect = 6e-14
Identities = 40/90 (44%), Positives = 57/90 (62%), Gaps = 2/90 (2%)
Frame = -2
Query: 464 QLRGAIDPKRHYKKGDSKSKTLPKYFQMGTVVDSPLDFFSGRLTKKERRASLADELLADQ 285
++R ++DPKR YKK D PKYFQ+GT+VD+P DF+ R+ KK+R+ ++ +ELLAD
Sbjct: 665 KMRASMDPKRFYKKNDRDG--FPKYFQIGTIVDNPADFYHSRIPKKQRKRTIVEELLADS 722
Query: 284 NLVAYRKRKVREI--EEQNKPAGNENWKIK 201
Y +RK EI E+ AG + K K
Sbjct: 723 EFRRYNRRKYSEIMAEKAANAAGKKFRKKK 752
>ref|NP_722501.1| hypothetical protein MGC27716 [Mus musculus]
gi|25048631|ref|XP_194118.1| expressed sequence AA408582
[Mus musculus] gi|20271421|gb|AAH28305.1| Similar to
acidic 82 kDa protein mRNA [Mus musculus]
Length = 758
Score = 77.8 bits (190), Expect = 8e-14
Identities = 39/88 (44%), Positives = 55/88 (62%)
Frame = -2
Query: 464 QLRGAIDPKRHYKKGDSKSKTLPKYFQMGTVVDSPLDFFSGRLTKKERRASLADELLADQ 285
++R +DPKR YKK D PKYFQ+GT+VD+P DF+ R+ KK+R+ ++ +ELLAD
Sbjct: 667 KMRAGMDPKRFYKKNDRDG--FPKYFQVGTIVDNPADFYHSRIPKKQRKKTIVEELLADS 724
Query: 284 NLVAYRKRKVREIEEQNKPAGNENWKIK 201
+ +RK EI + K A E K K
Sbjct: 725 EFRRFNRRKYSEIMAE-KAANAEGKKFK 751
>ref|XP_215688.1| similar to hypothetical protein MGC27716 [Mus musculus] [Rattus
norvegicus]
Length = 109
Score = 77.0 bits (188), Expect = 1e-13
Identities = 38/88 (43%), Positives = 56/88 (63%)
Frame = -2
Query: 464 QLRGAIDPKRHYKKGDSKSKTLPKYFQMGTVVDSPLDFFSGRLTKKERRASLADELLADQ 285
++R ++DPKR YKK D PKYFQ+GT+VD+P DF+ R+ KK+R+ ++ +ELLAD
Sbjct: 18 KMRASMDPKRFYKKNDRDG--FPKYFQVGTIVDNPADFYHSRIPKKQRKKTIVEELLADS 75
Query: 284 NLVAYRKRKVREIEEQNKPAGNENWKIK 201
+ +RK EI + K A E K +
Sbjct: 76 EFRRFNRRKYSEIMAE-KAANAEGKKFR 102
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 455,005,550
Number of Sequences: 1393205
Number of extensions: 9484143
Number of successful extensions: 25365
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 24777
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25336
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18947112822
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)