Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
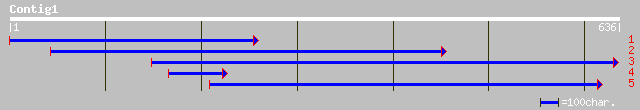
Query= KMC003270A_C01 KMC003270A_c01
(636 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_220668.1| NADH DEHYDROGENASE I CHAIN N (nuoN2) [Rickettsi... 36 0.49
ref|NP_360019.1| NADH dehydrogenase I chain N [EC:1.6.5.3] [Rick... 35 0.64
gb|EAA25570.1| NADH dehydrogenase I chain N [Rickettsia sibirica] 35 0.64
ref|NP_567967.1| E3 ubiquitin ligase SCF complex subunit SKP1/AS... 33 4.1
ref|NP_199205.1| WD-repeat protein-like; protein id: At5g43920.1... 33 4.1
>ref|NP_220668.1| NADH DEHYDROGENASE I CHAIN N (nuoN2) [Rickettsia prowazekii]
gi|7467895|pir||G71683 NADH2 dehydrogenase (ubiquinone)
(EC 1.6.5.3) I chain N2 RP284 - Rickettsia prowazekii
gi|3860845|emb|CAA14745.1| NADH DEHYDROGENASE I CHAIN N
(nuoN2) [Rickettsia prowazekii]
Length = 432
Score = 35.8 bits (81), Expect = 0.49
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Frame = -2
Query: 605 FKSIAEILKTVFIPYNAWVTRFVNNNHS-IFSHLHDAANFLNIPSLFEFTSLFTGLE 438
F I ILKT F P + W+ R NN S I +L + + I +++FT + G E
Sbjct: 220 FFLIGSILKTAFFPMHFWMMRAYNNTASIILVYLAGISTIIGIYIIYKFTYIIIGYE 276
>ref|NP_360019.1| NADH dehydrogenase I chain N [EC:1.6.5.3] [Rickettsia conorii]
gi|25285085|pir||F97747 NADH2 dehydrogenase (ubiquinone)
(EC 1.6.5.3) - Rickettsia conorii (strain Malish 7)
gi|15619448|gb|AAL02920.1| NADH dehydrogenase I chain N
[EC:1.6.5.3] [Rickettsia conorii]
Length = 433
Score = 35.4 bits (80), Expect = 0.64
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Frame = -2
Query: 605 FKSIAEILKTVFIPYNAWVTRFVNNNHS-IFSHLHDAANFLNIPSLFEFTSLFTGLE 438
F I ILKT F P + W+ R NN S I +L + + I +++FT + G E
Sbjct: 220 FFLIGAILKTAFFPMHFWMMRAYNNTASVILVYLAGISTIIGIYIIYKFTYIIIGYE 276
>gb|EAA25570.1| NADH dehydrogenase I chain N [Rickettsia sibirica]
Length = 371
Score = 35.4 bits (80), Expect = 0.64
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Frame = -2
Query: 605 FKSIAEILKTVFIPYNAWVTRFVNNNHS-IFSHLHDAANFLNIPSLFEFTSLFTGLE 438
F I ILKT F P + W+ R NN S I +L + + I +++FT + G E
Sbjct: 158 FFLIGAILKTAFFPMHFWMMRAYNNTASVILVYLAGISTIIGIYIIYKFTYIIIGYE 214
>ref|NP_567967.1| E3 ubiquitin ligase SCF complex subunit SKP1/ASK1 (At12), putative;
protein id: At4g34470.1 [Arabidopsis thaliana]
gi|7488340|pir||T05267 SKP1-like protein T4L20.50 -
Arabidopsis thaliana gi|3096916|emb|CAA18826.1|
Skp1p-like protein [Arabidopsis thaliana]
gi|7270397|emb|CAB80164.1| Skp1p-like protein
[Arabidopsis thaliana]
Length = 152
Score = 32.7 bits (73), Expect = 4.1
Identities = 18/53 (33%), Positives = 27/53 (49%)
Frame = -2
Query: 560 NAWVTRFVNNNHSIFSHLHDAANFLNIPSLFEFTSLFTGLEDRGENENKIQET 402
N W +F++ S L AAN+LNI SLF+ T +G+ +I+ T
Sbjct: 76 NKWDEKFMDLEQSTIFELILAANYLNIKSLFDLTCQTVADMIKGKTPEEIRST 128
>ref|NP_199205.1| WD-repeat protein-like; protein id: At5g43920.1 [Arabidopsis
thaliana] gi|9758551|dbj|BAB09052.1| WD-repeat
protein-like [Arabidopsis thaliana]
Length = 523
Score = 32.7 bits (73), Expect = 4.1
Identities = 15/22 (68%), Positives = 16/22 (72%)
Frame = +2
Query: 83 VNFGSWNPTNPHMLASASNGVT 148
VN SWNP NP MLASAS+ T
Sbjct: 488 VNCVSWNPKNPRMLASASDDQT 509
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 486,952,266
Number of Sequences: 1393205
Number of extensions: 9523516
Number of successful extensions: 19643
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 19185
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19639
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)