Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003228A_C01 KMC003228A_c01
(607 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
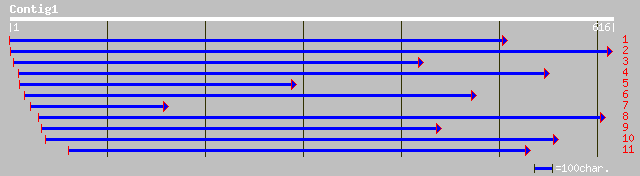
pir||F86203 hypothetical protein [imported] - Arabidopsis thalia... 118 5e-26
ref|NP_172172.2| unknown protein; protein id: At1g06890.1, suppo... 118 5e-26
gb|AAO00831.1| putative integral membrane protein [Arabidopsis t... 113 2e-24
ref|NP_180604.1| putative integral membrane protein; protein id:... 113 2e-24
dbj|BAB39904.1| contains ESTs D48306(S14443),D24269(R1613),AU076... 98 7e-20
>pir||F86203 hypothetical protein [imported] - Arabidopsis thaliana
gi|7523696|gb|AAF63135.1|AC011001_5 Hypothetical protein
[Arabidopsis thaliana]
Length = 255
Score = 118 bits (296), Expect = 5e-26
Identities = 55/77 (71%), Positives = 65/77 (83%)
Frame = -3
Query: 605 LVLAFGYILLRDPFSWRNILGILVAMIGMILYSYYCSLESQQKAVEAAAQASQAREAESD 426
LVLAFGY+LLRDPF WRNILGILVA+IGM++YSYYCS+E+QQKA E + Q Q +E+E D
Sbjct: 160 LVLAFGYVLLRDPFDWRNILGILVAVIGMVVYSYYCSIETQQKASETSTQLPQMKESEKD 219
Query: 425 PLISVENGSGVASDSVG 375
PLI+ ENGSGV SD G
Sbjct: 220 PLIAAENGSGVLSDGGG 236
>ref|NP_172172.2| unknown protein; protein id: At1g06890.1, supported by cDNA:
gi_19423945 [Arabidopsis thaliana]
gi|19423946|gb|AAL87295.1| unknown protein [Arabidopsis
thaliana] gi|21436373|gb|AAM51356.1| unknown protein
[Arabidopsis thaliana]
Length = 357
Score = 118 bits (296), Expect = 5e-26
Identities = 55/77 (71%), Positives = 65/77 (83%)
Frame = -3
Query: 605 LVLAFGYILLRDPFSWRNILGILVAMIGMILYSYYCSLESQQKAVEAAAQASQAREAESD 426
LVLAFGY+LLRDPF WRNILGILVA+IGM++YSYYCS+E+QQKA E + Q Q +E+E D
Sbjct: 262 LVLAFGYVLLRDPFDWRNILGILVAVIGMVVYSYYCSIETQQKASETSTQLPQMKESEKD 321
Query: 425 PLISVENGSGVASDSVG 375
PLI+ ENGSGV SD G
Sbjct: 322 PLIAAENGSGVLSDGGG 338
>gb|AAO00831.1| putative integral membrane protein [Arabidopsis thaliana]
Length = 353
Score = 113 bits (282), Expect = 2e-24
Identities = 53/75 (70%), Positives = 63/75 (83%)
Frame = -3
Query: 605 LVLAFGYILLRDPFSWRNILGILVAMIGMILYSYYCSLESQQKAVEAAAQASQAREAESD 426
LVLAFGY+LL+D FSWRNILGILVA+IGM+LYSYYC+LE+QQKA E + Q Q E E D
Sbjct: 262 LVLAFGYLLLKDAFSWRNILGILVAVIGMVLYSYYCTLETQQKATETSTQLPQMDENEKD 321
Query: 425 PLISVENGSGVASDS 381
PL+S ENGSG+ SD+
Sbjct: 322 PLVSAENGSGLISDN 336
>ref|NP_180604.1| putative integral membrane protein; protein id: At2g30460.1
[Arabidopsis thaliana] gi|25408076|pir||F84708 probable
integral membrane protein [imported] - Arabidopsis
thaliana gi|1946372|gb|AAB63090.1| putative integral
membrane protein [Arabidopsis thaliana]
Length = 200
Score = 113 bits (282), Expect = 2e-24
Identities = 53/75 (70%), Positives = 63/75 (83%)
Frame = -3
Query: 605 LVLAFGYILLRDPFSWRNILGILVAMIGMILYSYYCSLESQQKAVEAAAQASQAREAESD 426
LVLAFGY+LL+D FSWRNILGILVA+IGM+LYSYYC+LE+QQKA E + Q Q E E D
Sbjct: 109 LVLAFGYLLLKDAFSWRNILGILVAVIGMVLYSYYCTLETQQKATETSTQLPQMDENEKD 168
Query: 425 PLISVENGSGVASDS 381
PL+S ENGSG+ SD+
Sbjct: 169 PLVSAENGSGLISDN 183
>dbj|BAB39904.1| contains ESTs D48306(S14443),D24269(R1613),AU076096(E20048)~similar
to Arabidopsis thaliana chromosome 1, F4H5.5~unknown
protein [Oryza sativa (japonica cultivar-group)]
gi|15528768|dbj|BAB64810.1| P0701D05.25 [Oryza sativa
(japonica cultivar-group)] gi|20804811|dbj|BAB92494.1|
OJ1276_B06.3 [Oryza sativa (japonica cultivar-group)]
Length = 356
Score = 98.2 bits (243), Expect = 7e-20
Identities = 51/85 (60%), Positives = 62/85 (72%), Gaps = 5/85 (5%)
Frame = -3
Query: 605 LVLAFGYILLRDPFSWRNILGILVAMIGMILYSYYCSLESQQKAVEAAAQASQAREAESD 426
LVLAFGY+LL DPFSWRNILGIL+A+IGM+ YSY+C+ E+ K EA+ Q +Q +E+ESD
Sbjct: 263 LVLAFGYVLLHDPFSWRNILGILIAVIGMVSYSYFCTKEAPPKPTEASPQLNQVKESESD 322
Query: 425 PLIS-----VENGSGVASDSVGTKV 366
PLIS ENG G A D KV
Sbjct: 323 PLISDSLSTAENG-GNAGDDEALKV 346
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 525,316,373
Number of Sequences: 1393205
Number of extensions: 10859260
Number of successful extensions: 27235
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 26520
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27228
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)