Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003209A_C01 KMC003209A_c01
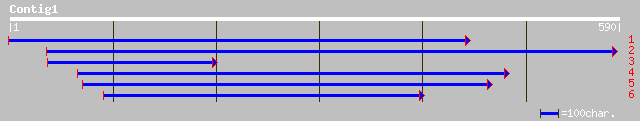
(585 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_172097.1| bZIP protein; protein id: At1g06070.1, supporte... 101 8e-21
gb|AAM62924.1| transcriptional activator RF2a, putative [Arabido... 101 8e-21
ref|NP_180695.1| bZIP transcription factor (POSF21); protein id:... 93 3e-18
gb|AAB05810.1| super cysteine rich protein; SCRP [Homo sapiens] 75 6e-13
gb|AAD01862.1| posF21 [Arabidopsis thaliana] 74 1e-12
>ref|NP_172097.1| bZIP protein; protein id: At1g06070.1, supported by cDNA: 17213.
[Arabidopsis thaliana] gi|25406894|pir||H86195
hypothetical protein [imported] - Arabidopsis thaliana
gi|8810469|gb|AAF80130.1|AC024174_12 Contains similarity
to a b-Zip binding protein from Arabidopsis thaliana
gb|Z86093 and contains a b-Zip transcription factor
PF|00170 domain. ESTs gb|AV551499, gb|T04752,
gb|AV550784, gb|AV550336, gb|AV545846, gb|AV538486,
gb|AV542369, gb|AV538179 come from this gene
gi|17065886|emb|CAD12037.1| AtbZIP transcription factor
[Arabidopsis thaliana]
Length = 423
Score = 101 bits (251), Expect = 8e-21
Identities = 55/88 (62%), Positives = 61/88 (68%), Gaps = 6/88 (6%)
Frame = -1
Query: 585 GPMMNYASFGGGQQFYPHNHSMHTLLAAQQFQQLQIHPQKHQQHQHQFQQQIQQQQQQ-- 412
G MNY SFG QQFYP+N SMHT+LAAQQ QQLQI QK QQ Q Q QQQ QQQQQQ
Sbjct: 312 GTSMNYGSFGSNQQFYPNNQSMHTILAAQQLQQLQIQSQKQQQQQQQHQQQQQQQQQQFH 371
Query: 411 -QQQ---QIQQQQQQMQQQQPGTTSTIR 340
QQQ Q+QQQQ+ QQ+Q S +R
Sbjct: 372 FQQQQLYQLQQQQRLQQQEQQSGASELR 399
>gb|AAM62924.1| transcriptional activator RF2a, putative [Arabidopsis thaliana]
Length = 423
Score = 101 bits (251), Expect = 8e-21
Identities = 55/88 (62%), Positives = 61/88 (68%), Gaps = 6/88 (6%)
Frame = -1
Query: 585 GPMMNYASFGGGQQFYPHNHSMHTLLAAQQFQQLQIHPQKHQQHQHQFQQQIQQQQQQ-- 412
G MNY SFG QQFYP+N SMHT+LAAQQ QQLQI QK QQ Q Q QQQ QQQQQQ
Sbjct: 312 GTSMNYGSFGSNQQFYPNNQSMHTILAAQQLQQLQIQSQKQQQQQQQHQQQQQQQQQQFH 371
Query: 411 -QQQ---QIQQQQQQMQQQQPGTTSTIR 340
QQQ Q+QQQQ+ QQ+Q S +R
Sbjct: 372 FQQQQLYQLQQQQRLQQQEQQSGASELR 399
>ref|NP_180695.1| bZIP transcription factor (POSF21); protein id: At2g31370.1,
supported by cDNA: gi_15982853 [Arabidopsis thaliana]
gi|1172441|sp|Q04088|PF21_ARATH Possible transcription
factor PosF21 gi|99685|pir||S21883 bZIP transcription
factor (POSF21) [imported] - Arabidopsis thaliana
gi|16429|emb|CAA43366.1| posF21 [Arabidopsis thaliana]
gi|4589968|gb|AAD26486.1| bZIP transcription factor
(POSF21) [Arabidopsis thaliana]
gi|15982854|gb|AAL09774.1| At2g31370/T28P16.14
[Arabidopsis thaliana] gi|21360501|gb|AAM47366.1|
At2g31370/T28P16.14 [Arabidopsis thaliana]
Length = 398
Score = 92.8 bits (229), Expect = 3e-18
Identities = 50/70 (71%), Positives = 54/70 (76%), Gaps = 1/70 (1%)
Frame = -1
Query: 576 MNYASFGGGQQ-FYPHNHSMHTLLAAQQFQQLQIHPQKHQQHQHQFQQQIQQQQQQQQQQ 400
+NY SFG QQ FY +N SM T+LAA+QFQQLQIH QK QQ Q Q QQQ QQQQQQQQQ
Sbjct: 304 LNYGSFGSNQQQFYSNNQSMQTILAAKQFQQLQIHSQKQQQQQQQ-QQQQHQQQQQQQQQ 362
Query: 399 IQQQQQQMQQ 370
Q QQQQMQQ
Sbjct: 363 YQFQQQQMQQ 372
>gb|AAB05810.1| super cysteine rich protein; SCRP [Homo sapiens]
Length = 46
Score = 75.1 bits (183), Expect = 6e-13
Identities = 21/25 (84%), Positives = 21/25 (84%)
Frame = +2
Query: 365 CCCCICCCCCCICCCCCCCCC*ICC 439
CCCC CCCCCC CCCCCCCCC CC
Sbjct: 12 CCCCCCCCCCCCCCCCCCCCCCFCC 36
Score = 74.7 bits (182), Expect = 8e-13
Identities = 21/25 (84%), Positives = 21/25 (84%)
Frame = +2
Query: 365 CCCCICCCCCCICCCCCCCCC*ICC 439
CCCC CCCCCC CCCCCCCCC CC
Sbjct: 13 CCCCCCCCCCCCCCCCCCCCCFCCC 37
Score = 73.2 bits (178), Expect = 2e-12
Identities = 23/32 (71%), Positives = 24/32 (74%)
Frame = +2
Query: 365 CCCCICCCCCCICCCCCCCCC*ICC*N*CWCC 460
CCCC CCCCCC CCCCCCCCC C C+CC
Sbjct: 11 CCCCCCCCCCCCCCCCCCCCC--C----CFCC 36
Score = 67.0 bits (162), Expect = 2e-10
Identities = 24/40 (60%), Positives = 25/40 (62%), Gaps = 5/40 (12%)
Frame = +2
Query: 362 GCCCC-----ICCCCCCICCCCCCCCC*ICC*N*CWCCWC 466
GC C CCCCCC CCCCCCCCC CC C CC+C
Sbjct: 1 GCELCPDRSRCCCCCCCCCCCCCCCCC--CC---CCCCFC 35
Score = 60.1 bits (144), Expect = 2e-08
Identities = 19/27 (70%), Positives = 19/27 (70%)
Frame = +2
Query: 389 CCCICCCCCCCCC*ICC*N*CWCCWCF 469
CCC CCCCCCCCC CC C CC CF
Sbjct: 11 CCCCCCCCCCCCCCCCC---CCCCCCF 34
Score = 35.4 bits (80), Expect = 0.53
Identities = 9/12 (75%), Positives = 10/12 (83%)
Frame = +2
Query: 365 CCCCICCCCCCI 400
CCCC CC CCC+
Sbjct: 27 CCCCCCCFCCCL 38
>gb|AAD01862.1| posF21 [Arabidopsis thaliana]
Length = 58
Score = 74.3 bits (181), Expect = 1e-12
Identities = 38/49 (77%), Positives = 40/49 (81%)
Frame = -1
Query: 516 TLLAAQQFQQLQIHPQKHQQHQHQFQQQIQQQQQQQQQQIQQQQQQMQQ 370
T+LAA+QFQQLQIH QK QQ Q Q QQQ QQQQQQQQQ Q QQQQMQQ
Sbjct: 2 TILAAKQFQQLQIHSQKQQQQQQQQQQQQHQQQQQQQQQYQFQQQQMQQ 50
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 460,312,428
Number of Sequences: 1393205
Number of extensions: 9988911
Number of successful extensions: 451400
Number of sequences better than 10.0: 6927
Number of HSP's better than 10.0 without gapping: 72261
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 209543
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)