Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003200A_C01 KMC003200A_c01
(641 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
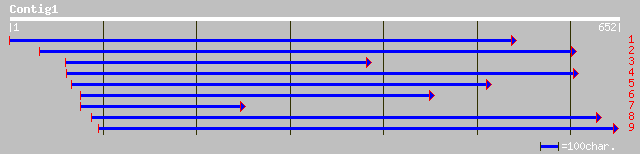
emb|CAD56219.1| ribosomal protein S3a [Cicer arietinum] 238 5e-62
sp|P33444|RS3A_CATRO 40S RIBOSOMAL PROTEIN S3A (CYC07 PROTEIN) 217 9e-56
ref|NP_195193.1| Putative S-phase-specific ribosomal protein; pr... 217 1e-55
dbj|BAA89498.1| cyc07 [Daucus carota] 216 2e-55
sp|P49198|RS3A_HELAN 40S RIBOSOMAL PROTEIN S3A gi|7441118|pir||T... 214 6e-55
>emb|CAD56219.1| ribosomal protein S3a [Cicer arietinum]
Length = 261
Score = 238 bits (607), Expect = 5e-62
Identities = 118/126 (93%), Positives = 122/126 (96%)
Frame = -3
Query: 639 RMFCIAFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDLKELVRKFIPEMIGK 460
RMFCI FTKRRSNQVKRTCYAQSSQIRQIRRKMREIM+NQATSCDLKELVRKFIPEMIGK
Sbjct: 136 RMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMVNQATSCDLKELVRKFIPEMIGK 195
Query: 459 EIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVGTKVDRPADETVVEGA 280
EIEKATSSIYPLQNVFIRKVKILK+PKFDLGKLMEVHGDYSEDVGTKV+ PADETVVEG
Sbjct: 196 EIEKATSSIYPLQNVFIRKVKILKSPKFDLGKLMEVHGDYSEDVGTKVESPADETVVEGT 255
Query: 279 TEVVGA 262
E+VGA
Sbjct: 256 PEIVGA 261
>sp|P33444|RS3A_CATRO 40S RIBOSOMAL PROTEIN S3A (CYC07 PROTEIN)
Length = 260
Score = 217 bits (553), Expect = 9e-56
Identities = 107/126 (84%), Positives = 118/126 (92%)
Frame = -3
Query: 639 RMFCIAFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDLKELVRKFIPEMIGK 460
RMFCI FTK+R+NQ KRTCYAQSSQIRQIRRKMREIM+NQA SCDLK+LV+KFIPE IG+
Sbjct: 136 RMFCIGFTKKRANQQKRTCYAQSSQIRQIRRKMREIMVNQAQSCDLKDLVQKFIPESIGR 195
Query: 459 EIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVGTKVDRPADETVVEGA 280
EIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY+ED+GTK+DRPA+E V E
Sbjct: 196 EIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYNEDIGTKLDRPAEEAVAE-P 254
Query: 279 TEVVGA 262
TEV+GA
Sbjct: 255 TEVIGA 260
>ref|NP_195193.1| Putative S-phase-specific ribosomal protein; protein id:
At4g34670.1, supported by cDNA: gi_17064847 [Arabidopsis
thaliana] gi|19884297|sp|Q42262|RS3A_ARATH 40S ribosomal
protein S3A gi|7441117|pir||T05287 ribosomal protein
S3a, cytosolic - Arabidopsis thaliana
gi|2661422|emb|CAA04689.1| Putative S-phase-specific
ribosomal protein [Arabidopsis thaliana]
gi|3096936|emb|CAA18846.1| Putative S-phase-specific
ribosomal protein [Arabidopsis thaliana]
gi|7270417|emb|CAB80184.1| Putative S-phase-specific
ribosomal protein [Arabidopsis thaliana]
gi|17064848|gb|AAL32578.1| Putative S-phase-specific
ribosomal protein [Arabidopsis thaliana]
Length = 262
Score = 217 bits (552), Expect = 1e-55
Identities = 108/127 (85%), Positives = 117/127 (92%), Gaps = 1/127 (0%)
Frame = -3
Query: 639 RMFCIAFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDLKELVRKFIPEMIGK 460
RMFCIAFTKRR+NQVKRTCYAQSSQIRQIRRKM EIM+ +A+SCDLKELV KFIPE IG+
Sbjct: 136 RMFCIAFTKRRANQVKRTCYAQSSQIRQIRRKMSEIMVKEASSCDLKELVAKFIPEAIGR 195
Query: 459 EIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY-SEDVGTKVDRPADETVVEG 283
EIEKAT IYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY +EDVG KVDRPADET+VE
Sbjct: 196 EIEKATQGIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYTAEDVGVKVDRPADETMVEE 255
Query: 282 ATEVVGA 262
TE++GA
Sbjct: 256 PTEIIGA 262
>dbj|BAA89498.1| cyc07 [Daucus carota]
Length = 261
Score = 216 bits (551), Expect = 2e-55
Identities = 107/126 (84%), Positives = 117/126 (91%)
Frame = -3
Query: 639 RMFCIAFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDLKELVRKFIPEMIGK 460
RMFCI FTK+R+NQ KRTCYAQSSQIRQIRRKM EIM NQA+SCDLKELV KFIPE IG+
Sbjct: 136 RMFCIGFTKKRANQQKRTCYAQSSQIRQIRRKMVEIMRNQASSCDLKELVAKFIPESIGR 195
Query: 459 EIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVGTKVDRPADETVVEGA 280
EIEKATSSI+PLQNVFIRKVKILKAPKFD+GKLMEVHGDYSEDVG K++RP +ET+VEG
Sbjct: 196 EIEKATSSIFPLQNVFIRKVKILKAPKFDIGKLMEVHGDYSEDVGVKLERPIEETMVEGE 255
Query: 279 TEVVGA 262
TEVVGA
Sbjct: 256 TEVVGA 261
>sp|P49198|RS3A_HELAN 40S RIBOSOMAL PROTEIN S3A gi|7441118|pir||T09301 ribosomal protein
S3a - common sunflower gi|469248|gb|AAA80978.1|
ribosomal protein S3a
Length = 260
Score = 214 bits (546), Expect = 6e-55
Identities = 106/126 (84%), Positives = 117/126 (92%)
Frame = -3
Query: 639 RMFCIAFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCDLKELVRKFIPEMIGK 460
RMFCI FTK+R+NQVKRTCYAQSSQI+QIRRKMREIM+ QA +CDLKELV+KFIPE IG+
Sbjct: 136 RMFCIGFTKKRANQVKRTCYAQSSQIKQIRRKMREIMVTQAQTCDLKELVQKFIPESIGR 195
Query: 459 EIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVGTKVDRPADETVVEGA 280
EIEKATSSIYPLQNVFIRKVKILKAPKFD+GKLMEVHGDYSEDVG K+DRPA+E E A
Sbjct: 196 EIEKATSSIYPLQNVFIRKVKILKAPKFDIGKLMEVHGDYSEDVGVKLDRPAEEAAPE-A 254
Query: 279 TEVVGA 262
TEV+GA
Sbjct: 255 TEVIGA 260
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 497,883,415
Number of Sequences: 1393205
Number of extensions: 10009690
Number of successful extensions: 28732
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 26412
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28516
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27007650415
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)