Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003194A_C01 KMC003194A_c01
(870 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
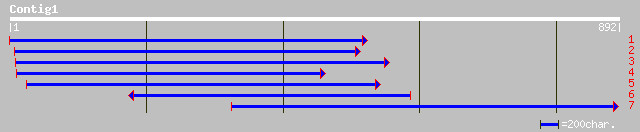
sp|P35016|ENPL_CATRO Endoplasmin homolog precursor (GRP94 homolo... 474 e-133
gb|AAL79732.1|AC091774_23 heat shock protein 90 [Oryza sativa] 468 e-131
gb|AAN34791.1| Grp94 [Xerophyta viscosa] 464 e-129
dbj|BAA90487.1| heat shock protein 90 [Oryza sativa] 461 e-129
dbj|BAB86368.1| SHEPHERD [Arabidopsis thaliana] 458 e-128
>sp|P35016|ENPL_CATRO Endoplasmin homolog precursor (GRP94 homolog) gi|542022|pir||S39558
HSP90 homolog - Madagascar periwinkle
gi|348696|gb|AAA16785.1| heat shock protein 90
Length = 817
Score = 474 bits (1219), Expect = e-133
Identities = 245/286 (85%), Positives = 258/286 (89%), Gaps = 5/286 (1%)
Frame = +1
Query: 28 MRKWTLPSALLLLSLLFLLCAD-----QGRKFQANAEGDSDELVDPPKVEDKIGAVPHGL 192
MRKWT+PS +LFLLC QGRK ANAE DSD VDPPKVEDKIGAVP+GL
Sbjct: 1 MRKWTVPS------VLFLLCPSLSSSCQGRKIHANAEADSDAPVDPPKVEDKIGAVPNGL 54
Query: 193 STDSDVAKREAESISKRSLRSNAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASD 372
STDSDVAKREAES+S R+LRS+AEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASD
Sbjct: 55 STDSDVAKREAESMSMRNLRSDAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASD 114
Query: 373 ALDKIRFLSLTDKEVLGEGDNTKLDIQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAK 552
ALDKIRFL+LTDKE+LGEGD KL+IQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAK
Sbjct: 115 ALDKIRFLALTDKEILGEGDTAKLEIQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAK 174
Query: 553 SGTSAFVEKMQTSGDLNLIGQLGVGFYSVYLVADYVEVISKHNDDNQYVWESKADGAFAI 732
SGTSAFVEKMQTSGDLNLIGQ GVGFYSVYLV DYVEVISKHNDD QY+WESKADGAFAI
Sbjct: 175 SGTSAFVEKMQTSGDLNLIGQFGVGFYSVYLVPDYVEVISKHNDDKQYIWESKADGAFAI 234
Query: 733 SEDTWNEPLGRGTEIRLHLKEEAGEYLEEFKLKEWVKRYSEFITFP 870
SED WNEPLGRGTEIRLHL++EA EYL+EFKLKE VKRYSEFI FP
Sbjct: 235 SEDVWNEPLGRGTEIRLHLRDEAQEYLDEFKLKELVKRYSEFINFP 280
>gb|AAL79732.1|AC091774_23 heat shock protein 90 [Oryza sativa]
Length = 812
Score = 468 bits (1204), Expect = e-131
Identities = 237/281 (84%), Positives = 256/281 (90%)
Frame = +1
Query: 28 MRKWTLPSALLLLSLLFLLCADQGRKFQANAEGDSDELVDPPKVEDKIGAVPHGLSTDSD 207
MRKW L SALLLL LL D +K Q NA+ +DELVDPPKVE+KIG VPHGLSTDS+
Sbjct: 1 MRKWALSSALLLLLLLLTTLPDPAKKLQVNADDSTDELVDPPKVEEKIGGVPHGLSTDSE 60
Query: 208 VAKREAESISKRSLRSNAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKI 387
V +REAESIS+++LRS+AEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKI
Sbjct: 61 VVQREAESISRKTLRSSAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKI 120
Query: 388 RFLSLTDKEVLGEGDNTKLDIQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSGTSA 567
RFL+LTDKEVLGEGD KL+IQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSGTSA
Sbjct: 121 RFLALTDKEVLGEGDTAKLEIQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSGTSA 180
Query: 568 FVEKMQTSGDLNLIGQLGVGFYSVYLVADYVEVISKHNDDNQYVWESKADGAFAISEDTW 747
FVEKMQT GDLNLIGQ GVGFYSVYLVADYVEVISKHNDD Q+VWESKADG+FAISEDTW
Sbjct: 181 FVEKMQTGGDLNLIGQFGVGFYSVYLVADYVEVISKHNDDKQHVWESKADGSFAISEDTW 240
Query: 748 NEPLGRGTEIRLHLKEEAGEYLEEFKLKEWVKRYSEFITFP 870
NEPLGRGTEIRLHL++EA EY+EE KLK+ VK+YSEFI FP
Sbjct: 241 NEPLGRGTEIRLHLRDEAKEYVEEDKLKDLVKKYSEFINFP 281
>gb|AAN34791.1| Grp94 [Xerophyta viscosa]
Length = 812
Score = 464 bits (1193), Expect = e-129
Identities = 241/284 (84%), Positives = 258/284 (89%), Gaps = 3/284 (1%)
Frame = +1
Query: 28 MRKWTLPSALLLLSLLFLLCA-DQGRKFQANAEG--DSDELVDPPKVEDKIGAVPHGLST 198
MR W++P AL+LL L+ L D GRK ANAE D+DELVDPPKVE+KI V GLST
Sbjct: 1 MRNWSIPPALVLLLLISLSAIPDGGRKLHANAEESRDADELVDPPKVEEKIAGVHGGLST 60
Query: 199 DSDVAKREAESISKRSLRSNAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDAL 378
D+DVAKREAES+S+++LRSNAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDAL
Sbjct: 61 DADVAKREAESMSRKNLRSNAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDAL 120
Query: 379 DKIRFLSLTDKEVLGEGDNTKLDIQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSG 558
DKIRFLSLTDKEVLGEGDNTKL+I IKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSG
Sbjct: 121 DKIRFLSLTDKEVLGEGDNTKLEIMIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSG 180
Query: 559 TSAFVEKMQTSGDLNLIGQLGVGFYSVYLVADYVEVISKHNDDNQYVWESKADGAFAISE 738
TSAFVEKMQT GDLNLIGQ GVGFYSVYLV+DYVEVISKHNDD QYVWESKADGAFAISE
Sbjct: 181 TSAFVEKMQTGGDLNLIGQFGVGFYSVYLVSDYVEVISKHNDDKQYVWESKADGAFAISE 240
Query: 739 DTWNEPLGRGTEIRLHLKEEAGEYLEEFKLKEWVKRYSEFITFP 870
DTWNEPLGRGTEIRLHL++EA EYL+E KLKE VK+YSEFI FP
Sbjct: 241 DTWNEPLGRGTEIRLHLRDEAKEYLDESKLKELVKKYSEFINFP 284
>dbj|BAA90487.1| heat shock protein 90 [Oryza sativa]
Length = 810
Score = 461 bits (1187), Expect = e-129
Identities = 236/281 (83%), Positives = 256/281 (90%)
Frame = +1
Query: 28 MRKWTLPSALLLLSLLFLLCADQGRKFQANAEGDSDELVDPPKVEDKIGAVPHGLSTDSD 207
MRKW L SALLLL L L D +K Q NA+ +DELVDPPKVE+KIG VPHGLSTDS+
Sbjct: 1 MRKWALSSALLLLLLTTL--PDPAKKLQVNADDSTDELVDPPKVEEKIGGVPHGLSTDSE 58
Query: 208 VAKREAESISKRSLRSNAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKI 387
V +REAESIS+++LRS+AEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKI
Sbjct: 59 VVQREAESISRKTLRSSAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKI 118
Query: 388 RFLSLTDKEVLGEGDNTKLDIQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSGTSA 567
RFL+LTDKEVLGEGD KL+IQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSGTSA
Sbjct: 119 RFLALTDKEVLGEGDTAKLEIQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSGTSA 178
Query: 568 FVEKMQTSGDLNLIGQLGVGFYSVYLVADYVEVISKHNDDNQYVWESKADGAFAISEDTW 747
FVEK+QT GDLNLIGQ GVGFYSVYLVADYVEVISKHNDD Q+VWESKADG+FAISEDTW
Sbjct: 179 FVEKVQTGGDLNLIGQFGVGFYSVYLVADYVEVISKHNDDKQHVWESKADGSFAISEDTW 238
Query: 748 NEPLGRGTEIRLHLKEEAGEYLEEFKLKEWVKRYSEFITFP 870
NEPLGRGTEIRLHL++EA EY+EE KLK+ VK+YSEFI FP
Sbjct: 239 NEPLGRGTEIRLHLRDEAKEYVEEDKLKDLVKKYSEFINFP 279
>dbj|BAB86368.1| SHEPHERD [Arabidopsis thaliana]
Length = 823
Score = 458 bits (1178), Expect = e-128
Identities = 238/281 (84%), Positives = 254/281 (89%)
Frame = +1
Query: 28 MRKWTLPSALLLLSLLFLLCADQGRKFQANAEGDSDELVDPPKVEDKIGAVPHGLSTDSD 207
MRK TL S L L SLLFLL DQGRK ANAE SD++ DPPKVE+KIG GLSTDSD
Sbjct: 1 MRKRTLVSVLFLFSLLFLL-PDQGRKLHANAEESSDDVTDPPKVEEKIGG-HGGLSTDSD 58
Query: 208 VAKREAESISKRSLRSNAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKI 387
V RE+ES+SK++LRSNAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKI
Sbjct: 59 VVHRESESMSKKTLRSNAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKI 118
Query: 388 RFLSLTDKEVLGEGDNTKLDIQIKLDKEKKILSIRDRGIGMTKEDLIKNLGTIAKSGTSA 567
RFL+LTDK+VLGEGD KL+IQIKLDK KKILSIRDRGIGMTKEDLIKNLGTIAKSGTSA
Sbjct: 119 RFLALTDKDVLGEGDTAKLEIQIKLDKAKKILSIRDRGIGMTKEDLIKNLGTIAKSGTSA 178
Query: 568 FVEKMQTSGDLNLIGQLGVGFYSVYLVADYVEVISKHNDDNQYVWESKADGAFAISEDTW 747
FVEKMQ+SGDLNLIGQ GVGFYS YLVADY+EVISKHNDD+QYVWESKADG FA+SEDTW
Sbjct: 179 FVEKMQSSGDLNLIGQFGVGFYSAYLVADYIEVISKHNDDSQYVWESKADGKFAVSEDTW 238
Query: 748 NEPLGRGTEIRLHLKEEAGEYLEEFKLKEWVKRYSEFITFP 870
NEPLGRGTEIRLHL++EAGEYLEE KLKE VKRYSEFI FP
Sbjct: 239 NEPLGRGTEIRLHLRDEAGEYLEESKLKELVKRYSEFINFP 279
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.315 0.135 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 749,906,454
Number of Sequences: 1393205
Number of extensions: 17157829
Number of successful extensions: 65491
Number of sequences better than 10.0: 540
Number of HSP's better than 10.0 without gapping: 60515
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 64274
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 46545945579
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)