Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003174A_C01 KMC003174A_c01
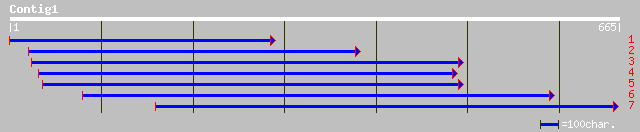
(665 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_563915.1| transport inhibitor response 1 (TIR1), putative... 205 5e-52
ref|NP_566800.1| transport inhibitor response 1 (TIR1), putative... 196 3e-49
emb|CAD40545.1| OSJNBa0072K14.1 [Oryza sativa (japonica cultivar... 177 1e-43
gb|AAG48832.1|AC084218_2 similar to Arabidopsis thaliana transpo... 166 3e-40
ref|NP_567135.1| transport inhibitor response 1 (TIR1), AtFBL1; ... 162 4e-39
>ref|NP_563915.1| transport inhibitor response 1 (TIR1), putative; protein id:
At1g12820.1, supported by cDNA: gi_20466150 [Arabidopsis
thaliana] gi|25352269|pir||F86261 F13K23.7 protein -
Arabidopsis thaliana gi|8698729|gb|AAF78487.1|AC012187_7
Strong similarity to transport inhibitor response 1
(TIR1) from Arabidopsis thaliana gb|AF005047
gi|20466151|gb|AAM20393.1| transport inhibitor response
1, putative [Arabidopsis thaliana]
gi|25083863|gb|AAN72129.1| transport inhibitor response
1, putative [Arabidopsis thaliana]
Length = 577
Score = 205 bits (521), Expect = 5e-52
Identities = 99/134 (73%), Positives = 113/134 (83%), Gaps = 3/134 (2%)
Frame = -2
Query: 664 TDQVFLYIGMYAEQLEMLSIAFAGESDKGMQYVLNGCKKLRKLEIRDCPFGNMALLADIG 485
TDQVFLYIGMYAEQLEMLSIAFAG++DKGM YVLNGCKK+RKLEIRD PFGN ALLAD+G
Sbjct: 441 TDQVFLYIGMYAEQLEMLSIAFAGDTDKGMLYVLNGCKKMRKLEIRDSPFGNAALLADVG 500
Query: 484 KYETMRSLWMSSCEVTVGACKELAEKMPRLNVEIFNENE---QEECSLEDEQSVEKMYLY 314
+YETMRSLWMSSCEVT+G CK LA+ PRLNVEI NENE E+ ++ + V+K+YLY
Sbjct: 501 RYETMRSLWMSSCEVTLGGCKRLAQNSPRLNVEIINENENNGMEQNEEDEREKVDKLYLY 560
Query: 313 RTLAGKRNDAPEYV 272
RT+ G R DAP YV
Sbjct: 561 RTVVGTRKDAPPYV 574
>ref|NP_566800.1| transport inhibitor response 1 (TIR1), putative; protein id:
At3g26810.1, supported by cDNA: gi_17064983 [Arabidopsis
thaliana] gi|9279671|dbj|BAB01228.1| transport inhibitor
response-like protein [Arabidopsis thaliana]
gi|17064984|gb|AAL32646.1| transport inhibitor
response-like protein [Arabidopsis thaliana]
Length = 575
Score = 196 bits (497), Expect = 3e-49
Identities = 98/137 (71%), Positives = 110/137 (79%), Gaps = 3/137 (2%)
Frame = -2
Query: 664 TDQVFLYIGMYAEQLEMLSIAFAGESDKGMQYVLNGCKKLRKLEIRDCPFGNMALLADIG 485
TDQVFLYIGMYA QLEMLSIAFAG++DKGM YVLNGCKK++KLEIRD PFG+ ALLAD+
Sbjct: 439 TDQVFLYIGMYANQLEMLSIAFAGDTDKGMLYVLNGCKKMKKLEIRDSPFGDTALLADVS 498
Query: 484 KYETMRSLWMSSCEVTVGACKELAEKMPRLNVEIFNENE---QEECSLEDEQSVEKMYLY 314
KYETMRSLWMSSCEVT+ CK LAEK P LNVEI NEN+ EE E Q V+K+YLY
Sbjct: 499 KYETMRSLWMSSCEVTLSGCKRLAEKAPWLNVEIINENDNNRMEENGHEGRQKVDKLYLY 558
Query: 313 RTLAGKRNDAPEYVCTL 263
RT+ G R DAP +V L
Sbjct: 559 RTVVGTRMDAPPFVWIL 575
>emb|CAD40545.1| OSJNBa0072K14.1 [Oryza sativa (japonica cultivar-group)]
Length = 575
Score = 177 bits (448), Expect = 1e-43
Identities = 89/135 (65%), Positives = 106/135 (77%), Gaps = 4/135 (2%)
Frame = -2
Query: 664 TDQVFLYIGMYAEQLEMLSIAFAGESDKGMQYVLNGCKKLRKLEIRDCPFGNMALLADIG 485
TD+VF+YIG YA+QLEMLSIAFAG+SDKGM +V+NGCK LRKLEIRD PFG+ ALL +
Sbjct: 439 TDKVFMYIGKYAKQLEMLSIAFAGDSDKGMMHVMNGCKNLRKLEIRDSPFGDAALLGNFA 498
Query: 484 KYETMRSLWMSSCEVTVGACKELAEKMPRLNVEIFNE----NEQEECSLEDEQSVEKMYL 317
+YETMRSLWMSSC VT+ C+ LA KMP LNVE+ NE NE EE + D VEK+Y+
Sbjct: 499 RYETMRSLWMSSCNVTLKGCQVLASKMPMLNVEVINERDGSNEMEE-NHGDLPKVEKLYV 557
Query: 316 YRTLAGKRNDAPEYV 272
YRT AG R+DAP +V
Sbjct: 558 YRTTAGARDDAPNFV 572
>gb|AAG48832.1|AC084218_2 similar to Arabidopsis thaliana transport inhibitor response 1
(TIR1) (T48087) [Oryza sativa]
Length = 587
Score = 166 bits (420), Expect = 3e-40
Identities = 84/134 (62%), Positives = 103/134 (76%), Gaps = 3/134 (2%)
Frame = -2
Query: 664 TDQVFLYIGMYAEQLEMLSIAFAGESDKGMQYVLNGCKKLRKLEIRDCPFGNMALLADIG 485
TD VF IG +A++LEMLSIAFAG SD G+ Y+L+GCK L+KLEIRDCPFG+ LLA+
Sbjct: 452 TDLVFKSIGAHADRLEMLSIAFAGNSDLGLHYILSGCKSLKKLEIRDCPFGDKPLLANAA 511
Query: 484 KYETMRSLWMSSCEVTVGACKELAEKMPRLNVEIFNENEQEEC---SLEDEQSVEKMYLY 314
K ETMRSLWMSSC +T+GAC++LA KMPRL+VEI N+ C SL DE VEK+Y+Y
Sbjct: 512 KLETMRSLWMSSCLLTLGACRQLARKMPRLSVEIMND-PGRSCPLDSLPDETPVEKLYVY 570
Query: 313 RTLAGKRNDAPEYV 272
RT+AG R+D P V
Sbjct: 571 RTIAGPRSDTPACV 584
>ref|NP_567135.1| transport inhibitor response 1 (TIR1), AtFBL1; protein id:
At3g62980.1, supported by cDNA: gi_11935208, supported
by cDNA: gi_2352493 [Arabidopsis thaliana]
gi|11282337|pir||T48087 transport inhibitor response
protein TIR1 [imported] - Arabidopsis thaliana
gi|2352492|gb|AAB69175.1| transport inhibitor response 1
[Arabidopsis thaliana] gi|2352494|gb|AAB69176.1|
transport inhibitor response 1 [Arabidopsis thaliana]
gi|7573427|emb|CAB87743.1| transport inhibitor response
1 (TIR1) [Arabidopsis thaliana]
gi|25054937|gb|AAN71945.1| putative transport inhibitor
response TIR1, AtFBL1 protein [Arabidopsis thaliana]
Length = 594
Score = 162 bits (410), Expect = 4e-39
Identities = 79/131 (60%), Positives = 102/131 (77%)
Frame = -2
Query: 664 TDQVFLYIGMYAEQLEMLSIAFAGESDKGMQYVLNGCKKLRKLEIRDCPFGNMALLADIG 485
TD+VF YIG YA+++EMLS+AFAG+SD GM +VL+GC LRKLEIRDCPFG+ ALLA+
Sbjct: 444 TDKVFEYIGTYAKKMEMLSVAFAGDSDLGMHHVLSGCDSLRKLEIRDCPFGDKALLANAS 503
Query: 484 KYETMRSLWMSSCEVTVGACKELAEKMPRLNVEIFNENEQEECSLEDEQSVEKMYLYRTL 305
K ETMRSLWMSSC V+ GACK L +KMP+LNVE+ +E + S + VE++++YRT+
Sbjct: 504 KLETMRSLWMSSCSVSFGACKLLGQKMPKLNVEVIDERGAPD-SRPESCPVERVFIYRTV 562
Query: 304 AGKRNDAPEYV 272
AG R D P +V
Sbjct: 563 AGPRFDMPGFV 573
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 587,225,095
Number of Sequences: 1393205
Number of extensions: 13034970
Number of successful extensions: 38130
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 36645
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38077
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28855580904
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)