Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003168A_C02 KMC003168A_c02
(543 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
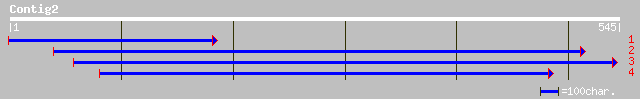
pir||T00410 protein kinase homolog T13E15.16 - Arabidopsis thali... 133 1e-30
ref|NP_182010.2| putative protein kinase; protein id: At2g44830.... 133 1e-30
gb|AAL84933.1| At2g44830/T13E15.16 [Arabidopsis thaliana] 133 1e-30
emb|CAC39054.1| putative protein kinase [Oryza sativa] 132 4e-30
gb|AAK31277.1|AC079890_13 putative protein kinase [Oryza sativa] 117 7e-26
>pir||T00410 protein kinase homolog T13E15.16 - Arabidopsis thaliana
gi|2344901|gb|AAC31841.1| putative protein kinase
[Arabidopsis thaliana]
Length = 762
Score = 133 bits (335), Expect = 1e-30
Identities = 68/103 (66%), Positives = 78/103 (75%), Gaps = 5/103 (4%)
Frame = -3
Query: 541 GRDLIRGLLVKEPQHRLGVKRGATEIKQHPFFEGVNWALIRCSTPPEVPRPVENEPPPPG 362
GRDLI+ LLVK+P++RLG KRGATEIKQHPFFEGVNWALIRCSTPPEVPR +E EPPP
Sbjct: 663 GRDLIQALLVKDPKNRLGTKRGATEIKQHPFFEGVNWALIRCSTPPEVPRQMETEPPP-- 720
Query: 361 KVGAVDNTVGIGSTSKRVVG-----GTTEMKPGGGRYLDFEFF 248
K G +D VG GS SKR++G GG++LDFEFF
Sbjct: 721 KYGPID-PVGFGSNSKRMMGPPAVSAAAADTKSGGKFLDFEFF 762
>ref|NP_182010.2| putative protein kinase; protein id: At2g44830.1, supported by
cDNA: gi_19310386 [Arabidopsis thaliana]
Length = 765
Score = 133 bits (335), Expect = 1e-30
Identities = 68/103 (66%), Positives = 78/103 (75%), Gaps = 5/103 (4%)
Frame = -3
Query: 541 GRDLIRGLLVKEPQHRLGVKRGATEIKQHPFFEGVNWALIRCSTPPEVPRPVENEPPPPG 362
GRDLI+ LLVK+P++RLG KRGATEIKQHPFFEGVNWALIRCSTPPEVPR +E EPPP
Sbjct: 666 GRDLIQALLVKDPKNRLGTKRGATEIKQHPFFEGVNWALIRCSTPPEVPRQMETEPPP-- 723
Query: 361 KVGAVDNTVGIGSTSKRVVG-----GTTEMKPGGGRYLDFEFF 248
K G +D VG GS SKR++G GG++LDFEFF
Sbjct: 724 KYGPID-PVGFGSNSKRMMGPPAVSAAAADTKSGGKFLDFEFF 765
>gb|AAL84933.1| At2g44830/T13E15.16 [Arabidopsis thaliana]
Length = 765
Score = 133 bits (335), Expect = 1e-30
Identities = 68/103 (66%), Positives = 78/103 (75%), Gaps = 5/103 (4%)
Frame = -3
Query: 541 GRDLIRGLLVKEPQHRLGVKRGATEIKQHPFFEGVNWALIRCSTPPEVPRPVENEPPPPG 362
GRDLI+ LLVK+P++RLG KRGATEIKQHPFFEGVNWALIRCSTPPEVPR +E EPPP
Sbjct: 666 GRDLIQALLVKDPKNRLGTKRGATEIKQHPFFEGVNWALIRCSTPPEVPRQMETEPPP-- 723
Query: 361 KVGAVDNTVGIGSTSKRVVG-----GTTEMKPGGGRYLDFEFF 248
K G +D VG GS SKR++G GG++LDFEFF
Sbjct: 724 KYGPID-PVGFGSNSKRMMGPPAVSAAAADTKSGGKFLDFEFF 765
>emb|CAC39054.1| putative protein kinase [Oryza sativa]
Length = 609
Score = 132 bits (331), Expect = 4e-30
Identities = 69/98 (70%), Positives = 71/98 (72%)
Frame = -3
Query: 541 GRDLIRGLLVKEPQHRLGVKRGATEIKQHPFFEGVNWALIRCSTPPEVPRPVENEPPPPG 362
GRDLIRGLLVKEPQ RLGVKRGA EIKQHPFFEGVNWALIRCSTPPEVPR VE E P
Sbjct: 517 GRDLIRGLLVKEPQQRLGVKRGAAEIKQHPFFEGVNWALIRCSTPPEVPRHVEAE--LPA 574
Query: 361 KVGAVDNTVGIGSTSKRVVGGTTEMKPGGGRYLDFEFF 248
K G + G KRVVG GG+YLDFEFF
Sbjct: 575 KYGVAEPVASGGGGGKRVVGAEVR---SGGKYLDFEFF 609
>gb|AAK31277.1|AC079890_13 putative protein kinase [Oryza sativa]
Length = 651
Score = 117 bits (294), Expect = 7e-26
Identities = 66/112 (58%), Positives = 77/112 (67%), Gaps = 14/112 (12%)
Frame = -3
Query: 541 GRDLIRGLLVKEPQHRLGVKRGATEIKQHPFFEGVNWALIRCSTPPEVPRPVE------- 383
GRDLIRGLL KEPQ RLGVKRGA EIKQHPFF+GVNWALIRCSTPP VPR VE
Sbjct: 542 GRDLIRGLLAKEPQGRLGVKRGAAEIKQHPFFDGVNWALIRCSTPPGVPRAVEPVAVAAA 601
Query: 382 ------NEPPPPGKVG-AVDNTVGIGSTSKRVVGGTTEMKPGGGRYLDFEFF 248
++PPP V A+ + ++SKR + G E++ GG+YLDFEFF
Sbjct: 602 VPATVMSKPPPVDTVEMAIHSNCNSTNSSKR-MAGPPEVE-SGGKYLDFEFF 651
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 488,019,315
Number of Sequences: 1393205
Number of extensions: 11205721
Number of successful extensions: 50696
Number of sequences better than 10.0: 794
Number of HSP's better than 10.0 without gapping: 41128
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 49841
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18750593680
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)