Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003160A_C01 KMC003160A_c01
(552 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
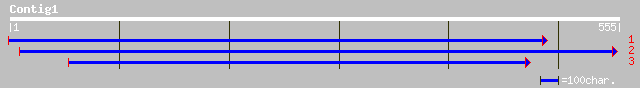
ref|NP_193103.1| hypothetical protein; protein id: At4g13670.1 [... 140 8e-33
ref|ZP_00118924.1| hypothetical protein [Cytophaga hutchinsonii] 43 0.002
ref|NP_181336.1| unknown protein; protein id: At2g38000.1, suppo... 41 0.008
ref|NP_190349.1| putative protein; protein id: At3g47650.1, supp... 40 0.014
dbj|BAB02057.1| gene_id:MKP6.23~unknown protein [Arabidopsis tha... 40 0.019
>ref|NP_193103.1| hypothetical protein; protein id: At4g13670.1 [Arabidopsis
thaliana] gi|7485696|pir||T05236 hypothetical protein
F18A5.60 - Arabidopsis thaliana
gi|4455296|emb|CAB36831.1| hypothetical protein
[Arabidopsis thaliana] gi|7268071|emb|CAB78409.1|
hypothetical protein [Arabidopsis thaliana]
Length = 432
Score = 140 bits (354), Expect = 8e-33
Identities = 65/111 (58%), Positives = 84/111 (75%), Gaps = 2/111 (1%)
Frame = -2
Query: 551 TEISDIQQKVVGG-GVKETEASRPRVFLLGENRWEDSSKL-GASDGVDRSKNKDGTTKCV 378
T++ + +Q +V +E + ++ RVFLLGENRWED S+L G + VDRS++ + T+C+
Sbjct: 318 TQVPEKKQSIVKDQSDREVDVTQNRVFLLGENRWEDPSRLIGRNKPVDRSESTNTKTRCI 377
Query: 377 QCRGEGRLLCTECDGGGEPNIEPQFMELVEDGTKCPYCEGLGYTPCDLCGG 225
CRGEGRL+C ECDG GEPNIEPQFME V + TKCPYCEGLGYT CD+C G
Sbjct: 378 TCRGEGRLMCLECDGTGEPNIEPQFMEWVGEDTKCPYCEGLGYTVCDVCDG 428
>ref|ZP_00118924.1| hypothetical protein [Cytophaga hutchinsonii]
Length = 211
Score = 43.1 bits (100), Expect = 0.002
Identities = 23/66 (34%), Positives = 31/66 (46%)
Frame = -2
Query: 422 GVDRSKNKDGTTKCVQCRGEGRLLCTECDGGGEPNIEPQFMELVEDGTKCPYCEGLGYTP 243
GV SK KC +C+G G+ C+ C G G + + + KC CEG G T
Sbjct: 130 GVGNSK-----VKCTKCKGVGKEYCSVCSGSG---VVISRSSMGDSYQKCYKCEGTGITL 181
Query: 242 CDLCGG 225
C +C G
Sbjct: 182 CSVCKG 187
Score = 35.8 bits (81), Expect = 0.35
Identities = 20/62 (32%), Positives = 27/62 (43%)
Frame = -2
Query: 434 GASDGVDRSKNKDGTTKCVQCRGEGRLLCTECDGGGEPNIEPQFMELVEDGTKCPYCEGL 255
G+ + RS D KC +C G G LC+ C G +DG CP C+G
Sbjct: 154 GSGVVISRSSMGDSYQKCYKCEGTGITLCSVCKG-----------TTFQDG-DCPQCKGH 201
Query: 254 GY 249
G+
Sbjct: 202 GF 203
>ref|NP_181336.1| unknown protein; protein id: At2g38000.1, supported by cDNA:
gi_19424072, supported by cDNA: gi_21281102 [Arabidopsis
thaliana] gi|25408585|pir||G84799 hypothetical protein
At2g38000 [imported] - Arabidopsis thaliana
gi|19424073|gb|AAL87350.1| unknown protein [Arabidopsis
thaliana] gi|21281103|gb|AAM45095.1| unknown protein
[Arabidopsis thaliana]
Length = 419
Score = 41.2 bits (95), Expect = 0.008
Identities = 24/72 (33%), Positives = 30/72 (41%), Gaps = 14/72 (19%)
Frame = -2
Query: 404 NKDGTTKCVQCRGEGRLLCTECDGGGEPNI--EPQFMELV----------EDG--TKCPY 267
N + KC C G G ++C C+ GEP E Q M+ +DG T C
Sbjct: 194 NSETVEKCTGCTGRGDVVCPTCNADGEPGFYKENQMMKCSTCYGRGLVAHKDGSDTICTN 253
Query: 266 CEGLGYTPCDLC 231
C G G PC C
Sbjct: 254 CNGKGKLPCPTC 265
>ref|NP_190349.1| putative protein; protein id: At3g47650.1, supported by cDNA:
gi_13877966, supported by cDNA: gi_17065647 [Arabidopsis
thaliana] gi|11280692|pir||T45725 hypothetical protein
F1P2.200 - Arabidopsis thaliana
gi|6522548|emb|CAB61991.1| putative protein [Arabidopsis
thaliana] gi|13877967|gb|AAK44061.1|AF370246_1 unknown
protein [Arabidopsis thaliana]
gi|17065648|gb|AAL33818.1| unknown protein [Arabidopsis
thaliana]
Length = 136
Score = 40.4 bits (93), Expect = 0.014
Identities = 20/59 (33%), Positives = 26/59 (43%)
Frame = -2
Query: 401 KDGTTKCVQCRGEGRLLCTECDGGGEPNIEPQFMELVEDGTKCPYCEGLGYTPCDLCGG 225
K + C C GEG + C++C GGG N+ F + G C C G C C G
Sbjct: 66 KPNSLVCANCEGEGCVACSQCKGGG-VNLIDHFNGQFKAGALCWLCRGKKEVLCGDCNG 123
>dbj|BAB02057.1| gene_id:MKP6.23~unknown protein [Arabidopsis thaliana]
Length = 135
Score = 40.0 bits (92), Expect = 0.019
Identities = 30/100 (30%), Positives = 48/100 (48%), Gaps = 2/100 (2%)
Frame = -2
Query: 518 GGGVKETEASRPRVFL-LGENRWEDSSKLGASDGVDRSKNKDGTTKCVQCRGEGRLLCTE 342
G GV +++++ + L L + W ++ GA + +SK K +CV C G GR+ C
Sbjct: 18 GEGVPNSKSTQGKTRLYLTKPSWIVRTQSGAKTCM-KSKAKG---RCVICHGSGRVDCFN 73
Query: 341 CDGGGEPN-IEPQFMELVEDGTKCPYCEGLGYTPCDLCGG 225
C G G N ++ + + E C C G G + C C G
Sbjct: 74 CCGKGRTNCVDVEMLPRGEWPKWCKSCGGSGLSDCSRCLG 113
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 521,653,576
Number of Sequences: 1393205
Number of extensions: 12533197
Number of successful extensions: 39397
Number of sequences better than 10.0: 124
Number of HSP's better than 10.0 without gapping: 37526
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39261
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19234190289
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)