Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
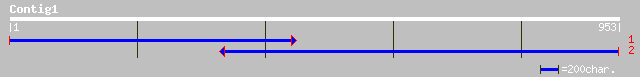
Query= KMC003152A_C01 KMC003152A_c01
(953 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_201260.1| putative protein; protein id: At5g64550.1, supp... 533 e-150
ref|NP_196529.1| putative protein; protein id: At5g09670.1 [Arab... 516 e-145
dbj|BAB21076.1| P0501G01.5 [Oryza sativa (japonica cultivar-group)] 482 e-135
pir||H96665 protein F22C12.10 [imported] - Arabidopsis thaliana ... 469 e-131
ref|NP_176596.1| hypothetical protein; protein id: At1g64140.1 [... 469 e-131
>ref|NP_201260.1| putative protein; protein id: At5g64550.1, supported by cDNA:
gi_20259337 [Arabidopsis thaliana]
gi|10178058|dbj|BAB11422.1|
emb|CAB89363.1~gene_id:MUB3.7~strong similarity to
unknown protein [Arabidopsis thaliana]
gi|20259338|gb|AAM13994.1| unknown protein [Arabidopsis
thaliana] gi|23296960|gb|AAN13211.1| unknown protein
[Arabidopsis thaliana]
Length = 634
Score = 533 bits (1374), Expect = e-150
Identities = 233/277 (84%), Positives = 257/277 (92%), Gaps = 1/277 (0%)
Frame = +2
Query: 125 TSENRISNPKRCRYFGCKKGARGASGLGIGHGGGQRCQKPGCNKGAESRTAYCKAHGGGR 304
+S+ R+SNPK+C++ GC KGARGASGL IGHGGGQRCQK GCNKGAES+T +CKAHGGG+
Sbjct: 205 SSQQRMSNPKKCKFMGCVKGARGASGLCIGHGGGQRCQKLGCNKGAESKTTFCKAHGGGK 264
Query: 305 RCNHLGCTKSAEGKTDYCIAHGGGKRCGYPDGCTKAARGKSGLCIRHGGGKRCRIEGCAR 484
RC HLGCTKSAEGKTD CI+HGGG+RCG+P+GC KAARGKSGLCI+HGGGKRCRIE C R
Sbjct: 265 RCQHLGCTKSAEGKTDLCISHGGGRRCGFPEGCAKAARGKSGLCIKHGGGKRCRIESCTR 324
Query: 485 SAEGQAGLCISHGGGRRCQYLGCSKGAQGSTMFCKAHGGGKRCSFAGCSKGAEGSTPLCK 664
SAEGQAGLCISHGGGRRCQ GC+KGAQGST +CKAHGGGKRC FAGC+KGAEGSTPLCK
Sbjct: 325 SAEGQAGLCISHGGGRRCQSSGCTKGAQGSTNYCKAHGGGKRCIFAGCTKGAEGSTPLCK 384
Query: 665 AHGGGKRCLYNGGGICGKSVHGGTNFCVAHGGGKRCAVSGCTKSARGRTDCCVRHGGGKR 844
AHGGGKRC+++GGGIC KSVHGGT+FCVAHGGGKRC V+GCTKSARGRTDCCV+HGGGKR
Sbjct: 385 AHGGGKRCMFDGGGICPKSVHGGTSFCVAHGGGKRCVVAGCTKSARGRTDCCVKHGGGKR 444
Query: 845 CKSEGCAKSAQGSTDFCKAHGGGKRCSW-GDGKCEKF 952
CKS+GC KSAQGSTDFCKAHGGGKRCSW GD KCEKF
Sbjct: 445 CKSDGCEKSAQGSTDFCKAHGGGKRCSWGGDWKCEKF 481
Score = 43.5 bits (101), Expect = 0.005
Identities = 19/50 (38%), Positives = 27/50 (54%)
Frame = +2
Query: 773 AVSGCTKSARGRTDCCVRHGGGKRCKSEGCAKSAQGSTDFCKAHGGGKRC 922
+VS + + T R K+CK GC K A+G++ C HGGG+RC
Sbjct: 192 SVSAFSDRSASATSSQQRMSNPKKCKFMGCVKGARGASGLCIGHGGGQRC 241
>ref|NP_196529.1| putative protein; protein id: At5g09670.1 [Arabidopsis thaliana]
gi|11357501|pir||T49931 hypothetical protein F17I14.140
- Arabidopsis thaliana gi|7671422|emb|CAB89363.1|
putative protein [Arabidopsis thaliana]
gi|9758995|dbj|BAB09522.1|
gb|AAF24563.1~gene_id:MTH16.9~strong similarity to
unknown protein [Arabidopsis thaliana]
gi|22530998|gb|AAM97003.1| putative protein [Arabidopsis
thaliana]
Length = 546
Score = 516 bits (1328), Expect = e-145
Identities = 226/275 (82%), Positives = 246/275 (89%), Gaps = 1/275 (0%)
Frame = +2
Query: 131 ENRISNPKRCRYFGCKKGARGASGLGIGHGGGQRCQKPGCNKGAESRTAYCKAHGGGRRC 310
+ R SNP++C++ GC KGARGASGL I HGGGQRCQKPGCNKGAES+T +CK HGGG+RC
Sbjct: 147 QQRTSNPRKCKFMGCVKGARGASGLCISHGGGQRCQKPGCNKGAESKTTFCKTHGGGKRC 206
Query: 311 NHLGCTKSAEGKTDYCIAHGGGKRCGYPDGCTKAARGKSGLCIRHGGGKRCRIEGCARSA 490
HLGCTKSAEGKTD+CI+HGGG+RC + +GC KAARG+SGLCI+HGGGKRC IE C RSA
Sbjct: 207 EHLGCTKSAEGKTDFCISHGGGRRCEFLEGCDKAARGRSGLCIKHGGGKRCNIEDCTRSA 266
Query: 491 EGQAGLCISHGGGRRCQYL-GCSKGAQGSTMFCKAHGGGKRCSFAGCSKGAEGSTPLCKA 667
EGQAGLCISHGGG+RCQY GC KGAQGST +CKAHGGGKRC F+GCSKGAEGSTPLCKA
Sbjct: 267 EGQAGLCISHGGGKRCQYFSGCEKGAQGSTNYCKAHGGGKRCIFSGCSKGAEGSTPLCKA 326
Query: 668 HGGGKRCLYNGGGICGKSVHGGTNFCVAHGGGKRCAVSGCTKSARGRTDCCVRHGGGKRC 847
HGGGKRCL +GGGIC KSVHGGTNFCVAHGGGKRC V GCTKSARGRTD CV+HGGGKRC
Sbjct: 327 HGGGKRCLADGGGICSKSVHGGTNFCVAHGGGKRCVVVGCTKSARGRTDSCVKHGGGKRC 386
Query: 848 KSEGCAKSAQGSTDFCKAHGGGKRCSWGDGKCEKF 952
K C KSAQGSTDFCKAHGGGKRCSWGDGKCEKF
Sbjct: 387 KIIDCEKSAQGSTDFCKAHGGGKRCSWGDGKCEKF 421
Score = 117 bits (294), Expect = 2e-25
Identities = 55/102 (53%), Positives = 63/102 (60%), Gaps = 4/102 (3%)
Frame = +2
Query: 152 KRCRYFG---CKKGARGASGLGIGHGGGQRCQKPGCNKGAESRTAYCKAHGGGRRCNHLG 322
KRC G C K G + + HGGG+RC GC K A RT C HGGG+RC +
Sbjct: 331 KRCLADGGGICSKSVHGGTNFCVAHGGGKRCVVVGCTKSARGRTDSCVKHGGGKRCKIID 390
Query: 323 CTKSAEGKTDYCIAHGGGKRCGYPDG-CTKAARGKSGLCIRH 445
C KSA+G TD+C AHGGGKRC + DG C K ARGKSGLC H
Sbjct: 391 CEKSAQGSTDFCKAHGGGKRCSWGDGKCEKFARGKSGLCAAH 432
>dbj|BAB21076.1| P0501G01.5 [Oryza sativa (japonica cultivar-group)]
Length = 646
Score = 482 bits (1240), Expect = e-135
Identities = 220/311 (70%), Positives = 248/311 (79%), Gaps = 12/311 (3%)
Frame = +2
Query: 56 AQSHPMLQ---ITHLALQSGLQDTGITSENRISNPKRCRYFGCKKGARGASGLGIGHGGG 226
A S P++ +T + I + R S K C++ GC KGARGASG I HGGG
Sbjct: 226 AMSSPVISSTLVTSMKSPVACTSGSINPQQRNSITKNCQFPGCVKGARGASGHCIAHGGG 285
Query: 227 QRCQKPGCNKGAESRTAYCKAHGGGRRCNHLGCTKSAEGKTDYCIAHGGGKRCGYPDGCT 406
+RCQKPGC KGAE RT YCKAHGGGRRC LGCTKSAEG+TD+CIAHGGG+RC + DGC+
Sbjct: 286 RRCQKPGCQKGAEGRTIYCKAHGGGRRCQFLGCTKSAEGRTDHCIAHGGGRRCSH-DGCS 344
Query: 407 KAARGKSGLCIRHGGGKRCRIEGCARSAEGQAGLCISHGGGRRCQYLGCSKGAQGSTMFC 586
+AARGKSGLCIRHGGGKRC+ E C RSAEG +G CISHGGGRRCQ+ C+KGAQGST FC
Sbjct: 345 RAARGKSGLCIRHGGGKRCQKENCIRSAEGHSGFCISHGGGRRCQFPECTKGAQGSTKFC 404
Query: 587 KAHGGGKRCSFAGCSKGAEGSTPLCKAHGGGKRCLYNGGGICGKSVHGGTNFCVAHGGGK 766
KAHGGGKRC+F+GC+KGAEGST CK HGGGKRCL+ GGG+C KSVHGGT +CVAHGGGK
Sbjct: 405 KAHGGGKRCTFSGCNKGAEGSTLFCKGHGGGKRCLFQGGGVCPKSVHGGTQYCVAHGGGK 464
Query: 767 RCAVSGCTKSARGRTDCCVRHGGGKRCKSEGCAKSAQGSTDFCKAHGGGKRCSWGD---- 934
RCA+SGCTKSARGRT+ CVRHGGGKRCK EGCAKSAQGSTDFCKAHGGGKRCSWG
Sbjct: 465 RCAISGCTKSARGRTEYCVRHGGGKRCKFEGCAKSAQGSTDFCKAHGGGKRCSWGQVDLN 524
Query: 935 -----GKCEKF 952
+C+KF
Sbjct: 525 FGVGAPQCDKF 535
Score = 217 bits (552), Expect = 2e-55
Identities = 99/186 (53%), Positives = 118/186 (63%), Gaps = 13/186 (6%)
Frame = +2
Query: 152 KRCRYFGCKKGARGASGLGIGHGGGQRCQKPGCNKGAESRTAYCKAHGGGRRCNHLGCTK 331
KRC+ C + A G SG I HGGG+RCQ P C KGA+ T +CKAHGGG+RC GC K
Sbjct: 361 KRCQKENCIRSAEGHSGFCISHGGGRRCQFPECTKGAQGSTKFCKAHGGGKRCTFSGCNK 420
Query: 332 SAEGKTDYCIAHGGGKRCGYPDG--CTKAARGKSGLCIRHGGGKRCRIEGCARSAEGQAG 505
AEG T +C HGGGKRC + G C K+ G + C+ HGGGKRC I GC +SA G+
Sbjct: 421 GAEGSTLFCKGHGGGKRCLFQGGGVCPKSVHGGTQYCVAHGGGKRCAISGCTKSARGRTE 480
Query: 506 LCISHGGGRRCQYLGCSKGAQGSTMFCKAHGGGKRCSFAG-----------CSKGAEGST 652
C+ HGGG+RC++ GC+K AQGST FCKAHGGGKRCS+ C K A T
Sbjct: 481 YCVRHGGGKRCKFEGCAKSAQGSTDFCKAHGGGKRCSWGQVDLNFGVGAPQCDKFARSKT 540
Query: 653 PLCKAH 670
LC AH
Sbjct: 541 GLCSAH 546
>pir||H96665 protein F22C12.10 [imported] - Arabidopsis thaliana
gi|6692098|gb|AAF24563.1|AC007764_5 F22C12.10
[Arabidopsis thaliana]
Length = 646
Score = 469 bits (1208), Expect = e-131
Identities = 212/289 (73%), Positives = 238/289 (81%), Gaps = 5/289 (1%)
Frame = +2
Query: 95 LQSGLQDTGITSE-----NRISNPKRCRYFGCKKGARGASGLGIGHGGGQRCQKPGCNKG 259
+ SG +G++ + S+ K C+ GC KGARGASG I HGGG+RCQK GC+KG
Sbjct: 233 ISSGTCTSGLSQQLKPQLKNSSSSKLCQVEGCHKGARGASGRCISHGGGRRCQKHGCHKG 292
Query: 260 AESRTAYCKAHGGGRRCNHLGCTKSAEGKTDYCIAHGGGKRCGYPDGCTKAARGKSGLCI 439
AE RT YCKAHGGGRRC LGCTKSAEG+TD+CIAHGGG+RC + D CT+AARG+SGLCI
Sbjct: 293 AEGRTVYCKAHGGGRRCEFLGCTKSAEGRTDFCIAHGGGRRCSHED-CTRAARGRSGLCI 351
Query: 440 RHGGGKRCRIEGCARSAEGQAGLCISHGGGRRCQYLGCSKGAQGSTMFCKAHGGGKRCSF 619
RHGGGKRC+ E C +SAEG +GLCISHGGGRRCQ GC+KGAQGSTMFCKAHGGGKRC+
Sbjct: 352 RHGGGKRCQRENCTKSAEGLSGLCISHGGGRRCQSNGCTKGAQGSTMFCKAHGGGKRCTH 411
Query: 620 AGCSKGAEGSTPLCKAHGGGKRCLYNGGGICGKSVHGGTNFCVAHGGGKRCAVSGCTKSA 799
+GC+KGAEGSTP CK HGGGKRC + G C KSVHGGTNFCVAHGGGKRCAV CTKSA
Sbjct: 412 SGCTKGAEGSTPFCKGHGGGKRCAFQGDDPCSKSVHGGTNFCVAHGGGKRCAVPECTKSA 471
Query: 800 RGRTDCCVRHGGGKRCKSEGCAKSAQGSTDFCKAHGGGKRCSWGDGKCE 946
RGRTD CVRHGGGKRC+SEGC KSAQGSTDFCKAHGGGKRC+WG + E
Sbjct: 472 RGRTDFCVRHGGGKRCQSEGCGKSAQGSTDFCKAHGGGKRCAWGQPETE 520
Score = 114 bits (285), Expect = 2e-24
Identities = 55/113 (48%), Positives = 65/113 (56%), Gaps = 15/113 (13%)
Frame = +2
Query: 152 KRCRYFG---CKKGARGASGLGIGHGGGQRCQKPGCNKGAESRTAYCKAHGGGRRCNHLG 322
KRC + G C K G + + HGGG+RC P C K A RT +C HGGG+RC G
Sbjct: 432 KRCAFQGDDPCSKSVHGGTNFCVAHGGGKRCAVPECTKSARGRTDFCVRHGGGKRCQSEG 491
Query: 323 CTKSAEGKTDYCIAHGGGKRC--GYPDG----------CTKAARGKSGLCIRH 445
C KSA+G TD+C AHGGGKRC G P+ CT ARGK+GLC H
Sbjct: 492 CGKSAQGSTDFCKAHGGGKRCAWGQPETEYAGQSSSGPCTSFARGKTGLCALH 544
>ref|NP_176596.1| hypothetical protein; protein id: At1g64140.1 [Arabidopsis thaliana]
Length = 658
Score = 469 bits (1208), Expect = e-131
Identities = 212/289 (73%), Positives = 238/289 (81%), Gaps = 5/289 (1%)
Frame = +2
Query: 95 LQSGLQDTGITSE-----NRISNPKRCRYFGCKKGARGASGLGIGHGGGQRCQKPGCNKG 259
+ SG +G++ + S+ K C+ GC KGARGASG I HGGG+RCQK GC+KG
Sbjct: 245 ISSGTCTSGLSQQLKPQLKNSSSSKLCQVEGCHKGARGASGRCISHGGGRRCQKHGCHKG 304
Query: 260 AESRTAYCKAHGGGRRCNHLGCTKSAEGKTDYCIAHGGGKRCGYPDGCTKAARGKSGLCI 439
AE RT YCKAHGGGRRC LGCTKSAEG+TD+CIAHGGG+RC + D CT+AARG+SGLCI
Sbjct: 305 AEGRTVYCKAHGGGRRCEFLGCTKSAEGRTDFCIAHGGGRRCSHED-CTRAARGRSGLCI 363
Query: 440 RHGGGKRCRIEGCARSAEGQAGLCISHGGGRRCQYLGCSKGAQGSTMFCKAHGGGKRCSF 619
RHGGGKRC+ E C +SAEG +GLCISHGGGRRCQ GC+KGAQGSTMFCKAHGGGKRC+
Sbjct: 364 RHGGGKRCQRENCTKSAEGLSGLCISHGGGRRCQSNGCTKGAQGSTMFCKAHGGGKRCTH 423
Query: 620 AGCSKGAEGSTPLCKAHGGGKRCLYNGGGICGKSVHGGTNFCVAHGGGKRCAVSGCTKSA 799
+GC+KGAEGSTP CK HGGGKRC + G C KSVHGGTNFCVAHGGGKRCAV CTKSA
Sbjct: 424 SGCTKGAEGSTPFCKGHGGGKRCAFQGDDPCSKSVHGGTNFCVAHGGGKRCAVPECTKSA 483
Query: 800 RGRTDCCVRHGGGKRCKSEGCAKSAQGSTDFCKAHGGGKRCSWGDGKCE 946
RGRTD CVRHGGGKRC+SEGC KSAQGSTDFCKAHGGGKRC+WG + E
Sbjct: 484 RGRTDFCVRHGGGKRCQSEGCGKSAQGSTDFCKAHGGGKRCAWGQPETE 532
Score = 114 bits (285), Expect = 2e-24
Identities = 55/113 (48%), Positives = 65/113 (56%), Gaps = 15/113 (13%)
Frame = +2
Query: 152 KRCRYFG---CKKGARGASGLGIGHGGGQRCQKPGCNKGAESRTAYCKAHGGGRRCNHLG 322
KRC + G C K G + + HGGG+RC P C K A RT +C HGGG+RC G
Sbjct: 444 KRCAFQGDDPCSKSVHGGTNFCVAHGGGKRCAVPECTKSARGRTDFCVRHGGGKRCQSEG 503
Query: 323 CTKSAEGKTDYCIAHGGGKRC--GYPDG----------CTKAARGKSGLCIRH 445
C KSA+G TD+C AHGGGKRC G P+ CT ARGK+GLC H
Sbjct: 504 CGKSAQGSTDFCKAHGGGKRCAWGQPETEYAGQSSSGPCTSFARGKTGLCALH 556
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 990,049,167
Number of Sequences: 1393205
Number of extensions: 28721171
Number of successful extensions: 133930
Number of sequences better than 10.0: 1646
Number of HSP's better than 10.0 without gapping: 90456
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 119474
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 53801056208
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)