Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003122A_C01 KMC003122A_c01
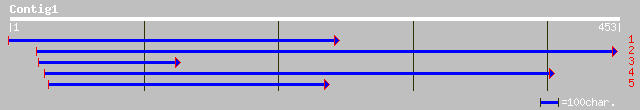
(423 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_175822.1| hypothetical protein; protein id: At1g54200.1 [... 102 8e-22
ref|NP_188014.1| hypothetical protein; protein id: At3g13980.1 [... 91 4e-18
ref|NP_189866.1| putative protein; protein id: At3g42800.1 [Arab... 58 2e-08
ref|NP_196766.1| putative serine rich protein; protein id: At5g1... 57 5e-08
gb|AAM65142.1| putative serine rich protein [Arabidopsis thaliana] 57 5e-08
>ref|NP_175822.1| hypothetical protein; protein id: At1g54200.1 [Arabidopsis
thaliana] gi|25372748|pir||C96583 hypothetical protein
F20D21.2 [imported] - Arabidopsis thaliana
gi|4585964|gb|AAD25600.1|AC005287_2 hypothetical protein
[Arabidopsis thaliana]
Length = 366
Score = 102 bits (255), Expect = 8e-22
Identities = 54/83 (65%), Positives = 65/83 (78%), Gaps = 2/83 (2%)
Frame = -3
Query: 367 VMNESRRVEELARELLKNYQKKNAG--DYDRMQCEDEDDDDAESCSSSDLFELDNLSAIG 194
VM E+RRV E A+ELL++YQKKN + E++DDDDA SC+SSDLFELDNLSAIG
Sbjct: 283 VMEENRRVIEAAKELLRSYQKKNKEVIEVSVEDDEEDDDDDALSCTSSDLFELDNLSAIG 342
Query: 193 IERYREELPVYETTPFQTHRAIA 125
I+RYREELPVYETT T+R I+
Sbjct: 343 IDRYREELPVYETTRLNTNRIIS 365
>ref|NP_188014.1| hypothetical protein; protein id: At3g13980.1 [Arabidopsis
thaliana] gi|11994369|dbj|BAB02328.1|
gb|AAD25600.1~gene_id:MDC16.10~similar to unknown
protein [Arabidopsis thaliana]
Length = 357
Score = 90.5 bits (223), Expect = 4e-18
Identities = 48/87 (55%), Positives = 62/87 (71%), Gaps = 2/87 (2%)
Frame = -3
Query: 364 MNESRRVEELARELLKNYQKKNAGDYDRMQ-CEDEDDDDAESCSSSDLFELDNLSAIGIE 188
+ E RRV + A++LL+ Y KN + E++D+DDA SC+SSDLFEL+NLSAIGIE
Sbjct: 271 VEEHRRVIQAAKDLLRTYHNKNKVTTTNINNVEEDDEDDAASCASSDLFELENLSAIGIE 330
Query: 187 RYREELPVYETTPFQT-HRAIAHGFVM 110
RYREELPVYETT +R IA G ++
Sbjct: 331 RYREELPVYETTRLDNMNRVIATGLIV 357
>ref|NP_189866.1| putative protein; protein id: At3g42800.1 [Arabidopsis thaliana]
gi|11358169|pir||T47338 hypothetical protein T21C14.20 -
Arabidopsis thaliana gi|7543888|emb|CAB87197.1| putative
protein [Arabidopsis thaliana]
Length = 341
Score = 58.2 bits (139), Expect = 2e-08
Identities = 30/52 (57%), Positives = 41/52 (78%)
Frame = -3
Query: 265 EDDDDAESCSSSDLFELDNLSAIGIERYREELPVYETTPFQTHRAIAHGFVM 110
E+++DA S SSSDLFELD+ IG+ RY +ELPVYETT F+T++AIA ++
Sbjct: 291 EEEEDAWSHSSSDLFELDSYR-IGMGRYLKELPVYETTDFKTNQAIARSLLL 341
>ref|NP_196766.1| putative serine rich protein; protein id: At5g12050.1, supported by
cDNA: 36958., supported by cDNA: gi_13877840, supported
by cDNA: gi_15983788, supported by cDNA: gi_16323505
[Arabidopsis thaliana] gi|11358674|pir||T48564 probable
serine rich protein - Arabidopsis thaliana
gi|7573372|emb|CAB87678.1| putative serine rich protein
[Arabidopsis thaliana]
gi|13877841|gb|AAK43998.1|AF370183_1 putative serine
rich protein [Arabidopsis thaliana]
gi|16323506|gb|AAL15247.1| putative serine rich protein
[Arabidopsis thaliana]
Length = 362
Score = 57.0 bits (136), Expect = 5e-08
Identities = 38/80 (47%), Positives = 46/80 (57%), Gaps = 13/80 (16%)
Frame = -3
Query: 352 RRVEELARELLKNYQKKNAG---------DYDRMQCEDEDDDDAESCSSSDLFELDNLSA 200
R VEE+ARE L++Y K + DY+ ED+DDDD S SSSDLFELD
Sbjct: 281 RSVEEIAREFLRDYHKNHENSLVKTNGFEDYEDDD-EDDDDDDVASDSSSDLFELD---L 336
Query: 199 IGIER----YREELPVYETT 152
+G Y +ELPVYETT
Sbjct: 337 VGNHHHHNVYGDELPVYETT 356
>gb|AAM65142.1| putative serine rich protein [Arabidopsis thaliana]
Length = 362
Score = 57.0 bits (136), Expect = 5e-08
Identities = 38/80 (47%), Positives = 46/80 (57%), Gaps = 13/80 (16%)
Frame = -3
Query: 352 RRVEELARELLKNYQKKNAG---------DYDRMQCEDEDDDDAESCSSSDLFELDNLSA 200
R VEE+ARE L++Y K + DY+ ED+DDDD S SSSDLFELD
Sbjct: 281 RSVEEIAREFLRDYHKNHENSLVKTNGFEDYEDDD-EDDDDDDVASDSSSDLFELD---L 336
Query: 199 IGIER----YREELPVYETT 152
+G Y +ELPVYETT
Sbjct: 337 VGNHHHHNVYGDELPVYETT 356
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 372,359,484
Number of Sequences: 1393205
Number of extensions: 8065024
Number of successful extensions: 41450
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 31353
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37548
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 6889859208
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)