Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
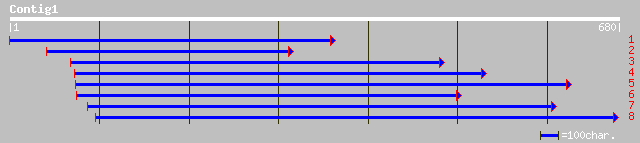
Query= KMC003120A_C01 KMC003120A_c01
(676 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_181084.1| chloroplast RNA-binding protein cp33, putative;... 105 6e-22
pir||T05725 cp31AHv protein - barley gi|3550467|emb|CAA06469.1| ... 67 3e-10
pir||S49463 RNA-binding protein RNP1 precursor - kidney bean gi|... 66 4e-10
pir||S23780 nucleic acid-binding protein - maize gi|168526|gb|AA... 66 4e-10
pir||T06232 Ps16 protein - wheat gi|2443390|dbj|BAA22411.1| Ps16... 65 6e-10
>ref|NP_181084.1| chloroplast RNA-binding protein cp33, putative; protein id:
At2g35410.1, supported by cDNA: gi_14596022 [Arabidopsis
thaliana] gi|25386549|pir||C84768 hypothetical protein
At2g35410 [imported] - Arabidopsis thaliana
gi|3608147|gb|AAC36180.1| putative chloroplast RNA
binding protein precursor [Arabidopsis thaliana]
gi|14596023|gb|AAK68739.1| putative chloroplast RNA
binding protein precursor [Arabidopsis thaliana]
gi|23198314|gb|AAN15684.1| putative chloroplast RNA
binding protein precursor [Arabidopsis thaliana]
Length = 308
Score = 105 bits (262), Expect = 6e-22
Identities = 56/117 (47%), Positives = 73/117 (61%), Gaps = 7/117 (5%)
Frame = -2
Query: 675 EARHVIYASNLAWKVRSTHLREFFTENFKAPLSARIVFDTPSGWATGYGFISYLTKEEAE 496
+ RH +Y SNLAWK RSTHLRE FT P+SAR+VF P G ++GYGF+S+ T+EEAE
Sbjct: 191 DTRHKLYVSNLAWKARSTHLRELFTAADFNPVSARVVFADPEGRSSGYGFVSFATREEAE 250
Query: 495 AAISALDGKELMGRSLFLKISEKKVKEA-------GSEKDEDLDQGHDAPTEES*IL 346
AI+ L+GKE+MGR + LK S + E+ + ED D D T E I+
Sbjct: 251 NAITKLNGKEIMGRPITLKFSLRSASESEDGDSVEANNASEDGDTVEDKNTSEENIV 307
>pir||T05725 cp31AHv protein - barley gi|3550467|emb|CAA06469.1| cp31AHv protein
[Hordeum vulgare subsp. vulgare]
Length = 294
Score = 66.6 bits (161), Expect = 3e-10
Identities = 31/79 (39%), Positives = 55/79 (69%)
Frame = -2
Query: 660 IYASNLAWKVRSTHLREFFTENFKAPLSARIVFDTPSGWATGYGFISYLTKEEAEAAISA 481
IY NL W+V + L E F+E+ K + A++V+D +G + G+GF++ ++EE + AI+A
Sbjct: 212 IYVGNLPWQVDDSRLVELFSEHGKV-VDAKVVYDRDTGRSRGFGFVTMASQEELDDAIAA 270
Query: 480 LDGKELMGRSLFLKISEKK 424
LDG+ L GR+L + ++E++
Sbjct: 271 LDGQSLEGRALRVNVAEER 289
Score = 37.4 bits (85), Expect = 0.19
Identities = 20/71 (28%), Positives = 37/71 (51%)
Frame = -2
Query: 660 IYASNLAWKVRSTHLREFFTENFKAPLSARIVFDTPSGWATGYGFISYLTKEEAEAAISA 481
+Y NL + V S L + F + +S ++++ + + G+GF++ T EEAE A+
Sbjct: 118 VYVGNLPYDVDSERLAQLFEQAGVVEVS-EVIYNRETDQSRGFGFVTMSTIEEAEKAVEM 176
Query: 480 LDGKELMGRSL 448
++ GR L
Sbjct: 177 FHRYDVNGRLL 187
>pir||S49463 RNA-binding protein RNP1 precursor - kidney bean
gi|558629|emb|CAA57551.1| chloroplast RNA binding
protein [Phaseolus vulgaris]
Length = 287
Score = 66.2 bits (160), Expect = 4e-10
Identities = 32/84 (38%), Positives = 53/84 (63%)
Frame = -2
Query: 675 EARHVIYASNLAWKVRSTHLREFFTENFKAPLSARIVFDTPSGWATGYGFISYLTKEEAE 496
E H ++ NL+W V + L + F E + + AR+++D +G + GYGF+ + TKEE E
Sbjct: 201 ETEHKLFVGNLSWSVTNEILTQAFQE-YGTVVGARVLYDGETGRSRGYGFVCFSTKEEME 259
Query: 495 AAISALDGKELMGRSLFLKISEKK 424
AA+ AL+ EL GR++ + ++E K
Sbjct: 260 AALGALNDVELEGRAMRVSLAEGK 283
Score = 45.8 bits (107), Expect = 5e-04
Identities = 28/83 (33%), Positives = 43/83 (51%), Gaps = 1/83 (1%)
Frame = -2
Query: 660 IYASNLAWKVRSTHLREFFTENFKAPLSARIVFDTPSGWATGYGFISYLTKEEAEAAISA 481
+Y NL + V S L + A L +++D +G + G+ F++ E+ A I
Sbjct: 115 LYFGNLPYSVDSAKLAGLIQDYGSAEL-IEVLYDRDTGKSRGFAFVTMSCIEDCNAVIEN 173
Query: 480 LDGKELMGRSLFLKISEK-KVKE 415
LDGKE +GR+L + S K K KE
Sbjct: 174 LDGKEYLGRTLRVNFSNKPKAKE 196
>pir||S23780 nucleic acid-binding protein - maize gi|168526|gb|AAA33486.1|
nucleic acid-binding protein
Length = 303
Score = 66.2 bits (160), Expect = 4e-10
Identities = 31/79 (39%), Positives = 55/79 (69%)
Frame = -2
Query: 660 IYASNLAWKVRSTHLREFFTENFKAPLSARIVFDTPSGWATGYGFISYLTKEEAEAAISA 481
IY NL W+V + L E F+E+ K + AR+V+D +G + G+GF++ +++E + AI+A
Sbjct: 221 IYVGNLPWQVDDSRLVELFSEHGKV-VDARVVYDRETGRSRGFGFVTMASQDELDDAIAA 279
Query: 480 LDGKELMGRSLFLKISEKK 424
LDG+ L GR+L + ++E++
Sbjct: 280 LDGQSLDGRALRVNVAEER 298
Score = 37.7 bits (86), Expect = 0.14
Identities = 20/71 (28%), Positives = 37/71 (51%)
Frame = -2
Query: 660 IYASNLAWKVRSTHLREFFTENFKAPLSARIVFDTPSGWATGYGFISYLTKEEAEAAISA 481
+Y NL + V S L + F + + A ++++ + + G+GF++ T EEAE A+
Sbjct: 127 VYVGNLPYDVDSERLAQLFDQAGVVEV-AEVIYNRETDQSRGFGFVTMSTVEEAEKAVEM 185
Query: 480 LDGKELMGRSL 448
++ GR L
Sbjct: 186 FHRYDVNGRLL 196
>pir||T06232 Ps16 protein - wheat gi|2443390|dbj|BAA22411.1| Ps16 protein
[Triticum aestivum]
Length = 293
Score = 65.5 bits (158), Expect = 6e-10
Identities = 31/79 (39%), Positives = 54/79 (68%)
Frame = -2
Query: 660 IYASNLAWKVRSTHLREFFTENFKAPLSARIVFDTPSGWATGYGFISYLTKEEAEAAISA 481
IY NL W+V + L E F+E+ K + AR+V+D +G + G+GF++ ++ E + AI+A
Sbjct: 211 IYVGNLPWQVDDSRLVELFSEHGKV-VDARVVYDRDTGRSRGFGFVTMASQPELDDAIAA 269
Query: 480 LDGKELMGRSLFLKISEKK 424
LDG+ L GR+L + ++E++
Sbjct: 270 LDGQSLEGRALRVNVAEER 288
Score = 37.4 bits (85), Expect = 0.19
Identities = 20/71 (28%), Positives = 37/71 (51%)
Frame = -2
Query: 660 IYASNLAWKVRSTHLREFFTENFKAPLSARIVFDTPSGWATGYGFISYLTKEEAEAAISA 481
+Y NL + V S L + F + +S ++++ + + G+GF++ T EEAE A+
Sbjct: 117 VYVGNLPYDVDSERLAQLFEQAGVVEVS-EVIYNRETDQSRGFGFVTMSTIEEAEKAVEM 175
Query: 480 LDGKELMGRSL 448
++ GR L
Sbjct: 176 FHRYDVNGRLL 186
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 574,487,428
Number of Sequences: 1393205
Number of extensions: 12270545
Number of successful extensions: 35182
Number of sequences better than 10.0: 914
Number of HSP's better than 10.0 without gapping: 33574
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35120
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29704274460
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)