Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003118A_C01 KMC003118A_c01
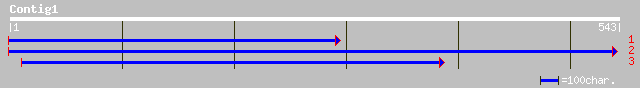
(541 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|EAA21526.1| rhoptry protein [Plasmodium yoelii yoelii] 37 0.12
ref|NP_504536.1| Serpentine Receptor, class D (delta) SRD-16 (sr... 37 0.20
pir||T25508 hypothetical protein C04E6.9 - Caenorhabditis elegans 37 0.20
gb|EAA16495.1| Ubiquitin carboxyl-terminal hydrolase family 2, p... 36 0.26
gb|EAA15918.1| asparagine-rich protein [Plasmodium yoelii yoelii] 35 0.58
>gb|EAA21526.1| rhoptry protein [Plasmodium yoelii yoelii]
Length = 2661
Score = 37.4 bits (85), Expect = 0.12
Identities = 29/148 (19%), Positives = 64/148 (42%), Gaps = 8/148 (5%)
Frame = +3
Query: 93 KNNKTEIKGSQNRIG--LNSKMKANTHLNNTTSKEASRDGKNSSISNIYALFPVPKCKRR 266
KNN T I N+ + S+ N +++ +K+ D N + + + ++
Sbjct: 2434 KNNDTSIISRINKFINLIKSEYNNNDNVSYNVAKKLEEDVNNIILDLDKSQNMLKDLIQQ 2493
Query: 267 NMHSVEDLRGEEEEIESDSHRAYTNRGKTIMNCYVINSS*YGKASKIEFVSQKXLIKFHE 446
N+ ++DL+ +++EIE+ ++ NR + I +N++ + ++ ++Q +
Sbjct: 2494 NLKIIDDLKNKKQEIENSNNLQTINREQEITQTEHVNNTYHHDINETNDINQNHQYSNSD 2553
Query: 447 KKSFIIVRANGKEIHQ------GTQNCY 512
KK + R G + G CY
Sbjct: 2554 KKEYSKTRDTGNSVRYAGAIAFGLVTCY 2581
>ref|NP_504536.1| Serpentine Receptor, class D (delta) SRD-16 (srd-16)
[Caenorhabditis elegans] gi|13775327|gb|AAK39148.1|
Serpentine receptor, class d (delta) protein 16
[Caenorhabditis elegans]
Length = 340
Score = 36.6 bits (83), Expect = 0.20
Identities = 23/53 (43%), Positives = 32/53 (59%), Gaps = 3/53 (5%)
Frame = -3
Query: 164 GVCFHFGV*ANSVLAALYLRFVILSRVGLLL---FLYLLSFLFSTKENNILLI 15
GVC HFG+ A +V +LYL + S LL+ + YL+ F + K NNILL+
Sbjct: 84 GVCKHFGLTACAVGFSLYLHTLTHSIWSLLISFAYRYLILFKTTFKRNNILLV 136
>pir||T25508 hypothetical protein C04E6.9 - Caenorhabditis elegans
Length = 308
Score = 36.6 bits (83), Expect = 0.20
Identities = 23/53 (43%), Positives = 32/53 (59%), Gaps = 3/53 (5%)
Frame = -3
Query: 164 GVCFHFGV*ANSVLAALYLRFVILSRVGLLL---FLYLLSFLFSTKENNILLI 15
GVC HFG+ A +V +LYL + S LL+ + YL+ F + K NNILL+
Sbjct: 84 GVCKHFGLTACAVGFSLYLHTLTHSIWSLLISFAYRYLILFKTTFKRNNILLV 136
>gb|EAA16495.1| Ubiquitin carboxyl-terminal hydrolase family 2, putative
[Plasmodium yoelii yoelii]
Length = 1172
Score = 36.2 bits (82), Expect = 0.26
Identities = 21/52 (40%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Frame = +3
Query: 78 KTNPAKNNKTEIKGSQNRIGLNSKMKANTHLNNTTSKE--ASRDGKNSSISN 227
+ N +KNN T+ GS+N NS + NT+ NN E +RD KNS N
Sbjct: 357 QNNGSKNNATQNNGSKNNSNQNSGGQNNTNKNNGDQNEDDKNRDDKNSDNQN 408
>gb|EAA15918.1| asparagine-rich protein [Plasmodium yoelii yoelii]
Length = 1238
Score = 35.0 bits (79), Expect = 0.58
Identities = 29/97 (29%), Positives = 43/97 (43%), Gaps = 1/97 (1%)
Frame = +3
Query: 84 NPAKNN-KTEIKGSQNRIGLNSKMKANTHLNNTTSKEASRDGKNSSISNIYALFPVPKCK 260
N KN+ K K S N G N N NN + + GKNS SNIY L K
Sbjct: 719 NSGKNSGKNSGKNSGNNGGNNG---GNNGGNNGGNNGGNNGGKNSGSSNIYEL------K 769
Query: 261 RRNMHSVEDLRGEEEEIESDSHRAYTNRGKTIMNCYV 371
+ N+++ + E EE + + + + KT + Y+
Sbjct: 770 KLNLYAENKKKKENEENDEKTFKNNIKKNKTKVFFYI 806
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 433,776,249
Number of Sequences: 1393205
Number of extensions: 9040043
Number of successful extensions: 34565
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 29056
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33543
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)