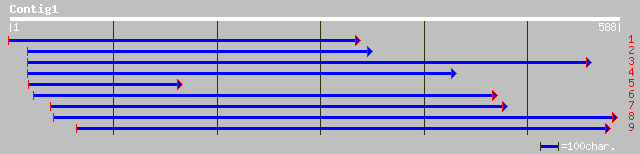
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003117A_C01 KMC003117A_c01
(563 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pdb|1H1Y|A Chain A, The Structure Of The Cytosolic D-Ribulose-5-... 146 9e-35
gb|AAF03437.1|AC010797_13 putative D-ribulose-5-phosphate 3-epim... 139 3e-34
ref|NP_566153.1| putative D-ribulose-5-phosphate 3-epimerase; pr... 139 3e-34
ref|NP_176518.1| putative D-ribulose-5-phosphate-3-epimerase; pr... 140 6e-34
gb|EAA34485.1| hypothetical protein [Neurospora crassa] 114 6e-25
>pdb|1H1Y|A Chain A, The Structure Of The Cytosolic D-Ribulose-5-Phosphate
3-Epimerase From Rice Complexed With Sulfate
gi|28373311|pdb|1H1Y|B Chain B, The Structure Of The
Cytosolic D-Ribulose-5-Phosphate 3-Epimerase From Rice
Complexed With Sulfate gi|28373312|pdb|1H1Z|A Chain A,
The Structure Of The Cytosolic D-Ribulose-5-Phosphate
3-Epimerase From Rice Complexed With Sulfate And Zinc
gi|28373313|pdb|1H1Z|B Chain B, The Structure Of The
Cytosolic D-Ribulose-5-Phosphate 3-Epimerase From Rice
Complexed With Sulfate And Zinc
gi|6007803|gb|AAF01048.1|AF189365_1
D-ribulose-5-phosphate 3-epimerase [Oryza sativa]
Length = 228
Score = 146 bits (369), Expect(2) = 9e-35
Identities = 71/79 (89%), Positives = 76/79 (95%)
Frame = -2
Query: 562 LVEAENPVEMVLVMTVAPGFGGQKFMPEMMDKVRILRKKYPSLDIEVDGGLGPSTIDMAA 383
LVEAENPVE+VLVMTV PGFGGQKFMPEMM+KVR LRKKYPSLDIEVDGGLGPSTID+AA
Sbjct: 132 LVEAENPVELVLVMTVEPGFGGQKFMPEMMEKVRALRKKYPSLDIEVDGGLGPSTIDVAA 191
Query: 382 SAGANCIVAGSSVFGAPDP 326
SAGANCIVAGSS+FGA +P
Sbjct: 192 SAGANCIVAGSSIFGAAEP 210
Score = 21.9 bits (45), Expect(2) = 9e-35
Identities = 10/16 (62%), Positives = 11/16 (68%)
Frame = -3
Query: 327 PAQVISLLRNSVEKGQ 280
P +VIS LR SVE Q
Sbjct: 210 PGEVISALRKSVEGSQ 225
>gb|AAF03437.1|AC010797_13 putative D-ribulose-5-phosphate 3-epimerase [Arabidopsis thaliana]
Length = 233
Score = 139 bits (351), Expect(2) = 3e-34
Identities = 67/79 (84%), Positives = 73/79 (91%)
Frame = -2
Query: 562 LVEAENPVEMVLVMTVAPGFGGQKFMPEMMDKVRILRKKYPSLDIEVDGGLGPSTIDMAA 383
LVE PVEMVLVMTV PGFGGQKFMP+MMDKVR LR+KYP+LDI+VDGGLGPSTID AA
Sbjct: 137 LVEGTTPVEMVLVMTVEPGFGGQKFMPDMMDKVRALRQKYPTLDIQVDGGLGPSTIDTAA 196
Query: 382 SAGANCIVAGSSVFGAPDP 326
+AGANCIVAGSSVFGAP+P
Sbjct: 197 AAGANCIVAGSSVFGAPEP 215
Score = 27.3 bits (59), Expect(2) = 3e-34
Identities = 13/18 (72%), Positives = 13/18 (72%)
Frame = -3
Query: 327 PAQVISLLRNSVEKGQQT 274
P VISLLR SVEK Q T
Sbjct: 215 PGDVISLLRTSVEKAQPT 232
>ref|NP_566153.1| putative D-ribulose-5-phosphate 3-epimerase; protein id:
At3g01850.1, supported by cDNA: 2398., supported by
cDNA: gi_13877556 [Arabidopsis thaliana]
gi|13877557|gb|AAK43856.1|AF370479_1 putative
D-ribulose-5-phosphate 3-epimerase [Arabidopsis
thaliana] gi|21592447|gb|AAM64398.1| putative
D-ribulose-5-phosphate 3-epimerase [Arabidopsis
thaliana] gi|25084145|gb|AAN72185.1| putative
D-ribulose-5-phosphate 3-epimerase [Arabidopsis
thaliana]
Length = 225
Score = 139 bits (351), Expect(2) = 3e-34
Identities = 67/79 (84%), Positives = 73/79 (91%)
Frame = -2
Query: 562 LVEAENPVEMVLVMTVAPGFGGQKFMPEMMDKVRILRKKYPSLDIEVDGGLGPSTIDMAA 383
LVE PVEMVLVMTV PGFGGQKFMP+MMDKVR LR+KYP+LDI+VDGGLGPSTID AA
Sbjct: 129 LVEGTTPVEMVLVMTVEPGFGGQKFMPDMMDKVRALRQKYPTLDIQVDGGLGPSTIDTAA 188
Query: 382 SAGANCIVAGSSVFGAPDP 326
+AGANCIVAGSSVFGAP+P
Sbjct: 189 AAGANCIVAGSSVFGAPEP 207
Score = 27.3 bits (59), Expect(2) = 3e-34
Identities = 13/18 (72%), Positives = 13/18 (72%)
Frame = -3
Query: 327 PAQVISLLRNSVEKGQQT 274
P VISLLR SVEK Q T
Sbjct: 207 PGDVISLLRTSVEKAQPT 224
>ref|NP_176518.1| putative D-ribulose-5-phosphate-3-epimerase; protein id:
At1g63290.1, supported by cDNA: 37843., supported by
cDNA: gi_20856481 [Arabidopsis thaliana]
gi|25292408|pir||G96658 probable D-ribulose-5-phosphate
F9N12.9 [imported] - Arabidopsis thaliana
gi|12324354|gb|AAG52146.1|AC022355_7 putative
D-ribulose-5-phosphate; 35237-36732 [Arabidopsis
thaliana] gi|20856482|gb|AAM26668.1| At1g63290/F9N12_9
[Arabidopsis thaliana] gi|21593298|gb|AAM65247.1|
putative D-ribulose-5-phosphate [Arabidopsis thaliana]
gi|23505807|gb|AAN28763.1| At1g63290/F9N12_9
[Arabidopsis thaliana]
Length = 227
Score = 140 bits (354), Expect(2) = 6e-34
Identities = 69/79 (87%), Positives = 71/79 (89%)
Frame = -2
Query: 562 LVEAENPVEMVLVMTVAPGFGGQKFMPEMMDKVRILRKKYPSLDIEVDGGLGPSTIDMAA 383
LVE NPVEMVLVMTV PGFGGQKFMP MMDKVR LR KYP+LDIEVDGGLGPSTID AA
Sbjct: 131 LVEGTNPVEMVLVMTVEPGFGGQKFMPSMMDKVRALRNKYPTLDIEVDGGLGPSTIDAAA 190
Query: 382 SAGANCIVAGSSVFGAPDP 326
+AGANCIVAGSSVFGAP P
Sbjct: 191 AAGANCIVAGSSVFGAPKP 209
Score = 25.0 bits (53), Expect(2) = 6e-34
Identities = 12/16 (75%), Positives = 12/16 (75%)
Frame = -3
Query: 327 PAQVISLLRNSVEKGQ 280
P VISLLR SVEK Q
Sbjct: 209 PGDVISLLRASVEKAQ 224
>gb|EAA34485.1| hypothetical protein [Neurospora crassa]
Length = 224
Score = 114 bits (286), Expect = 6e-25
Identities = 60/91 (65%), Positives = 70/91 (75%), Gaps = 3/91 (3%)
Frame = -2
Query: 562 LVEAENPVE---MVLVMTVAPGFGGQKFMPEMMDKVRILRKKYPSLDIEVDGGLGPSTID 392
++E+E+P E MVL+MTV PGFGGQKFM + KV+ LRK+YP L+IEVDGGLGPSTID
Sbjct: 125 ILESEDPKERPDMVLIMTVEPGFGGQKFMASELPKVQELRKRYPELNIEVDGGLGPSTID 184
Query: 391 MAASAGANCIVAGSSVFGAPDPSSSNFLTEE 299
AA AGAN IVAGS+VFGA DPS L E
Sbjct: 185 QAADAGANVIVAGSAVFGAQDPSEVIALLRE 215
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 466,671,564
Number of Sequences: 1393205
Number of extensions: 9747362
Number of successful extensions: 26960
Number of sequences better than 10.0: 216
Number of HSP's better than 10.0 without gapping: 25862
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26780
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20382500157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)