Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003102A_C01 KMC003102A_c01
(416 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
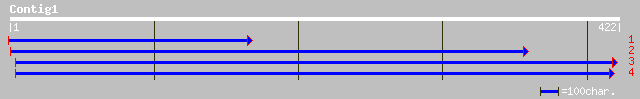
ref|NP_192233.1| putative frataxin-like protein; protein id: At4... 116 7e-26
pdb|1EKG|A Chain A, Mature Human Frataxin 79 2e-14
sp|Q16595|FRDA_HUMAN Frataxin, mitochondrial precursor (Friedrei... 79 2e-14
pdb|1LY7|A Chain A, The Solution Structure Of The The C-Terminal... 79 2e-14
ref|NP_000135.1| frataxin; Friedreich ataxia (frataxin) [Homo sa... 78 3e-14
>ref|NP_192233.1| putative frataxin-like protein; protein id: At4g03240.1
[Arabidopsis thaliana] gi|9910703|sp|Q9ZR07|FRDA_ARATH
Frataxin homolog gi|25407176|pir||B85041 probable
frataxin-like protein [imported] - Arabidopsis thaliana
gi|4262152|gb|AAD14452.1| putative frataxin-like protein
[Arabidopsis thaliana] gi|7270194|emb|CAB77809.1|
putative frataxin-like protein [Arabidopsis thaliana]
Length = 143
Score = 116 bits (290), Expect = 7e-26
Identities = 51/66 (77%), Positives = 58/66 (87%)
Frame = -3
Query: 414 VKLGDLGTYVLNKQTPNRQIWLSSPVSGPSRFDWDQDTKAWIYRRKKVNLYKLLEAELEQ 235
+KLG LGTYVLNKQTPNRQIW+SSPVSGPSRFDWD+D AWIYRR + L+KLLE ELE
Sbjct: 75 LKLGSLGTYVLNKQTPNRQIWMSSPVSGPSRFDWDRDANAWIYRRTEAKLHKLLEEELEN 134
Query: 234 LCGKPL 217
LCG+P+
Sbjct: 135 LCGEPI 140
>pdb|1EKG|A Chain A, Mature Human Frataxin
Length = 127
Score = 78.6 bits (192), Expect = 2e-14
Identities = 39/59 (66%), Positives = 46/59 (77%), Gaps = 1/59 (1%)
Frame = -3
Query: 414 VKLG-DLGTYVLNKQTPNRQIWLSSPVSGPSRFDWDQDTKAWIYRRKKVNLYKLLEAEL 241
VKLG DLGTYV+NKQTPN+QIWLSSP SGP R+DW K W+Y V+L++LL AEL
Sbjct: 51 VKLGGDLGTYVINKQTPNKQIWLSSPSSGPKRYDW--TGKNWVYSHDGVSLHELLAAEL 107
>sp|Q16595|FRDA_HUMAN Frataxin, mitochondrial precursor (Friedreich's ataxia protein)
(Fxn) gi|29127013|gb|AAH48097.1| similar to Friedreich
ataxia [Homo sapiens]
Length = 210
Score = 78.6 bits (192), Expect = 2e-14
Identities = 39/59 (66%), Positives = 46/59 (77%), Gaps = 1/59 (1%)
Frame = -3
Query: 414 VKLG-DLGTYVLNKQTPNRQIWLSSPVSGPSRFDWDQDTKAWIYRRKKVNLYKLLEAEL 241
VKLG DLGTYV+NKQTPN+QIWLSSP SGP R+DW K W+Y V+L++LL AEL
Sbjct: 134 VKLGGDLGTYVINKQTPNKQIWLSSPSSGPKRYDW--TGKNWVYSHDGVSLHELLAAEL 190
>pdb|1LY7|A Chain A, The Solution Structure Of The The C-Terminal Domain Of
Frataxin, The Protein Responsible For Friedreich Ataxia
Length = 121
Score = 78.6 bits (192), Expect = 2e-14
Identities = 39/59 (66%), Positives = 46/59 (77%), Gaps = 1/59 (1%)
Frame = -3
Query: 414 VKLG-DLGTYVLNKQTPNRQIWLSSPVSGPSRFDWDQDTKAWIYRRKKVNLYKLLEAEL 241
VKLG DLGTYV+NKQTPN+QIWLSSP SGP R+DW K W+Y V+L++LL AEL
Sbjct: 45 VKLGGDLGTYVINKQTPNKQIWLSSPSSGPKRYDW--TGKNWVYSHDGVSLHELLAAEL 101
>ref|NP_000135.1| frataxin; Friedreich ataxia (frataxin) [Homo sapiens]
gi|1218024|gb|AAA98508.1| frataxin
gi|1237439|gb|AAA98510.1| frataxin
Length = 210
Score = 77.8 bits (190), Expect = 3e-14
Identities = 43/76 (56%), Positives = 53/76 (69%), Gaps = 2/76 (2%)
Frame = -3
Query: 414 VKLG-DLGTYVLNKQTPNRQIWLSSPVSGPSRFDWDQDTKAWIYRRKKVNLYKLLEAELE 238
VKLG DLGTYV+NKQTPN+QIWLSSP SGP R+DW K W++ V+L++LL AEL
Sbjct: 134 VKLGGDLGTYVINKQTPNKQIWLSSPSSGPKRYDW--TGKNWVFSHDGVSLHELLAAELT 191
Query: 237 QLCGKPL-CSFLRY*G 193
+ L S+L Y G
Sbjct: 192 KALKTKLDLSWLAYSG 207
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 340,881,786
Number of Sequences: 1393205
Number of extensions: 6566128
Number of successful extensions: 13540
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 13318
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 13528
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 6956733048
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)