Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
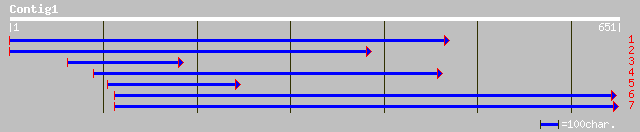
Query= KMC003086A_C01 KMC003086A_c01
(643 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL68832.1|AF463407_3 Enod8.1 [Medicago truncatula] 73 3e-12
pir||T52338 early nodule-specific protein ENOD8 [imported] - bar... 73 3e-12
pir||S59943 early nodulin 8 precursor - alfalfa gi|304037|gb|AAB... 72 8e-12
gb|AAA91034.1| nodulin 71 1e-11
dbj|BAB01482.1| gene_id:K24A2.4~similar to nodulins and lipase [... 70 2e-11
>gb|AAL68832.1|AF463407_3 Enod8.1 [Medicago truncatula]
Length = 381
Score = 73.2 bits (178), Expect = 3e-12
Identities = 35/74 (47%), Positives = 50/74 (67%), Gaps = 3/74 (4%)
Frame = -3
Query: 635 CCGNYLGGYKIS--CGQNTTESGT-VSDKTCKNPSEFLSWDGVHYSEEANLLIAKQILSG 465
CCG Y G Y I CG + +GT + +CKNPS + WDGVHY+E AN ++ QIL+G
Sbjct: 307 CCG-YGGEYNIGVGCGASININGTKIVAGSCKNPSTRIIWDGVHYTEAANEIVFSQILTG 365
Query: 464 AFSDPPVTIRQACF 423
F+DPP+++ +AC+
Sbjct: 366 VFNDPPISLDRACY 379
>pir||T52338 early nodule-specific protein ENOD8 [imported] - barrel medic
gi|3328240|gb|AAC26810.1| early nodule-specific protein
[Medicago truncatula]
Length = 381
Score = 73.2 bits (178), Expect = 3e-12
Identities = 35/74 (47%), Positives = 50/74 (67%), Gaps = 3/74 (4%)
Frame = -3
Query: 635 CCGNYLGGYKIS--CGQNTTESGT-VSDKTCKNPSEFLSWDGVHYSEEANLLIAKQILSG 465
CCG Y G Y I CG + +GT + +CKNPS + WDGVHY+E AN ++ QIL+G
Sbjct: 307 CCG-YGGEYNIGVGCGASININGTKIVAGSCKNPSTRIIWDGVHYTEAANEIVFSQILTG 365
Query: 464 AFSDPPVTIRQACF 423
F+DPP+++ +AC+
Sbjct: 366 VFNDPPISLDRACY 379
>pir||S59943 early nodulin 8 precursor - alfalfa gi|304037|gb|AAB41547.1| early
nodulin [Medicago sativa]
Length = 381
Score = 71.6 bits (174), Expect = 8e-12
Identities = 34/74 (45%), Positives = 48/74 (63%), Gaps = 3/74 (4%)
Frame = -3
Query: 635 CCGNYLGGYKIS--CGQNTTESGT-VSDKTCKNPSEFLSWDGVHYSEEANLLIAKQILSG 465
CCG Y G Y I CG +GT + +CKNPS ++WDG HY+E AN ++ QI +G
Sbjct: 307 CCG-YGGEYNIGAGCGATINVNGTKIVAGSCKNPSTRITWDGTHYTEAANKIVFDQISTG 365
Query: 464 AFSDPPVTIRQACF 423
AF+DPP+++ AC+
Sbjct: 366 AFNDPPISLDMACY 379
>gb|AAA91034.1| nodulin
Length = 381
Score = 71.2 bits (173), Expect = 1e-11
Identities = 34/74 (45%), Positives = 47/74 (62%), Gaps = 3/74 (4%)
Frame = -3
Query: 635 CCGNYLGGYKIS--CGQNTTESGT-VSDKTCKNPSEFLSWDGVHYSEEANLLIAKQILSG 465
CCG Y G Y I CG +GT + +CKNPS ++WDG HY+EEAN + QI +G
Sbjct: 307 CCG-YGGEYNIGAGCGATINVNGTKIVAGSCKNPSTRITWDGTHYTEEANKFVFYQISTG 365
Query: 464 AFSDPPVTIRQACF 423
F+DPP+++ AC+
Sbjct: 366 VFNDPPISLDMACY 379
>dbj|BAB01482.1| gene_id:K24A2.4~similar to nodulins and lipase [Arabidopsis
thaliana]
Length = 371
Score = 70.1 bits (170), Expect = 2e-11
Identities = 30/73 (41%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Frame = -3
Query: 641 EFCCGNYLGGYKISCGQNTTESGT-VSDKTCKNPSEFLSWDGVHYSEEANLLIAKQILSG 465
++CC +G + CG+ +GT + +C+N F+SWDG+HY+E AN+L+A +IL G
Sbjct: 295 DYCCVGAIGR-GMGCGKTIFLNGTELYSSSCQNRKNFISWDGIHYTETANMLVANRILDG 353
Query: 464 AFSDPPVTIRQAC 426
+ SDPP+ ++AC
Sbjct: 354 SISDPPLPTQKAC 366
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 542,026,239
Number of Sequences: 1393205
Number of extensions: 10928836
Number of successful extensions: 29484
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 28696
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29478
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27007650415
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)