Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003083A_C01 KMC003083A_c01
(514 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
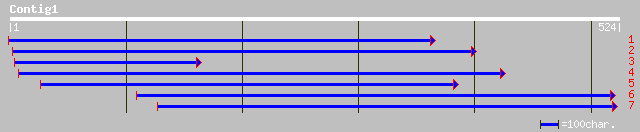
ref|NP_191256.1| bHLH protein; protein id: At3g56970.1, supporte... 48 8e-05
ref|NP_191257.1| bHLH protein; protein id: At3g56980.1, supporte... 44 0.001
ref|NP_181657.1| bHLH protein; protein id: At2g41240.1, supporte... 42 0.003
dbj|BAB64701.1| P0431G06.12 [Oryza sativa (japonica cultivar-gro... 38 0.079
ref|NP_196035.1| myc - like protein; protein id: At5g04150.1 [Ar... 37 0.13
>ref|NP_191256.1| bHLH protein; protein id: At3g56970.1, supported by cDNA:
gi_20127033 [Arabidopsis thaliana]
gi|11282360|pir||T47757 hypothetical protein F24I3.50 -
Arabidopsis thaliana gi|6911867|emb|CAB72167.1| putative
protein [Arabidopsis thaliana]
gi|20127034|gb|AAM10940.1|AF488576_1 putative bHLH
transcription factor [Arabidopsis thaliana]
Length = 253
Score = 47.8 bits (112), Expect = 8e-05
Identities = 26/77 (33%), Positives = 47/77 (60%), Gaps = 2/77 (2%)
Frame = -2
Query: 507 IHISSCEVYMTPLSEILLYLENNGHFLLNASSSETFEGRVFYNLHFQVEKA--HRIESCI 334
+ +SS +++ +S +L +E +G L++ SSS + R+FY LH QVE ++I
Sbjct: 177 VQVSSSKIHNFSISNVLGGIEEDGFVLVDVSSSRSQGERLFYTLHLQVENMDDYKINCEE 236
Query: 333 LTEKLLSIYEKKQSISN 283
L+E++L +YEK ++ N
Sbjct: 237 LSERMLYLYEKCENSFN 253
>ref|NP_191257.1| bHLH protein; protein id: At3g56980.1, supported by cDNA:
gi_20127035 [Arabidopsis thaliana]
gi|11282361|pir||T47758 hypothetical protein F24I3.60 -
Arabidopsis thaliana gi|6911868|emb|CAB72168.1| putative
protein [Arabidopsis thaliana]
gi|20127036|gb|AAM10941.1|AF488577_1 putative bHLH
transcription factor [Arabidopsis thaliana]
Length = 258
Score = 43.9 bits (102), Expect = 0.001
Identities = 24/71 (33%), Positives = 44/71 (61%), Gaps = 2/71 (2%)
Frame = -2
Query: 507 IHISSCEVYMTPLSEILLYLENNGHFLLNASSSETFEGRVFYNLHFQVEKA--HRIESCI 334
+ ISS +++ +S +L LE + L++ SSS + R+FY LH QVEK +++
Sbjct: 182 VQISSSKIHNFSISNVLSGLEEDRFVLVDMSSSRSQGERLFYTLHLQVEKIENYKLNCEE 241
Query: 333 LTEKLLSIYEK 301
L++++L +YE+
Sbjct: 242 LSQRMLYLYEE 252
>ref|NP_181657.1| bHLH protein; protein id: At2g41240.1, supported by cDNA:
gi_18491252 [Arabidopsis thaliana]
gi|25352412|pir||E84839 hypothetical protein At2g41240
[imported] - Arabidopsis thaliana
gi|3894198|gb|AAC78547.1| hypothetical protein
[Arabidopsis thaliana] gi|18491253|gb|AAL69451.1|
At2g41240/F13H10.21 [Arabidopsis thaliana]
Length = 242
Score = 42.4 bits (98), Expect = 0.003
Identities = 24/69 (34%), Positives = 40/69 (57%)
Frame = -2
Query: 507 IHISSCEVYMTPLSEILLYLENNGHFLLNASSSETFEGRVFYNLHFQVEKAHRIESCILT 328
+ ISS + +L +E +G L+ ASSS + R+FY++H Q+ K ++ S L
Sbjct: 169 VQISSLQTEKCSFGNVLSGVEEDGLVLVGASSSRSHGERLFYSMHLQI-KNGQVNSEELG 227
Query: 327 EKLLSIYEK 301
++LL +YEK
Sbjct: 228 DRLLYLYEK 236
>dbj|BAB64701.1| P0431G06.12 [Oryza sativa (japonica cultivar-group)]
Length = 248
Score = 37.7 bits (86), Expect = 0.079
Identities = 20/63 (31%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Frame = -2
Query: 474 PLSEILLYLENNGHFLLNASSSETFEGRVFYNLHFQVEKAHRIESC-ILTEKLLSIYEKK 298
PLS+ + LEN G +++S+S F R FY++H Q + E C E+L + K
Sbjct: 186 PLSKCIKVLENEGLHFISSSTSSGFGNRTFYSIHLQRSEGTINEECPAFCERLEKVVRNK 245
Query: 297 QSI 289
+
Sbjct: 246 AKL 248
>ref|NP_196035.1| myc - like protein; protein id: At5g04150.1 [Arabidopsis thaliana]
gi|9955570|emb|CAC05497.1| myc-like protein [Arabidopsis
thaliana] gi|26185711|emb|CAD58594.1| putative bHLH
transcription factor [Arabidopsis thaliana]
Length = 240
Score = 37.0 bits (84), Expect = 0.13
Identities = 17/38 (44%), Positives = 26/38 (67%)
Frame = -2
Query: 477 TPLSEILLYLENNGHFLLNASSSETFEGRVFYNLHFQV 364
T +S++LL LE NG +++ SSS + R+FY LH Q+
Sbjct: 181 TSVSDMLLRLEENGLNVISVSSSVSSTARIFYTLHLQM 218
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 396,975,956
Number of Sequences: 1393205
Number of extensions: 7526775
Number of successful extensions: 18089
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 17762
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18088
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 16232377112
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)