Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003080A_C01 KMC003080A_c01
(828 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
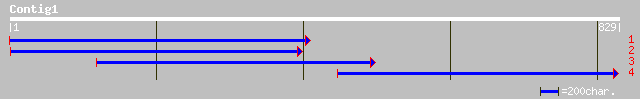
gb|AAL47685.1| p-coumarate 3-hydroxylase [Pinus taeda] 304 e-103
emb|CAD20576.1| putative cytochrome P450 [Solenostemon scutellar... 300 e-100
dbj|BAC44836.1| cytochrome P-450 [Lithospermum erythrorhizon] 285 1e-96
sp|O48956|C981_SORBI Cytochrome P450 98A1 gi|7430624|pir||T14638... 278 1e-95
sp|O48922|C982_SOYBN Cytochrome P450 98A2 gi|7430683|pir||T05937... 343 1e-93
>gb|AAL47685.1| p-coumarate 3-hydroxylase [Pinus taeda]
Length = 512
Score = 304 bits (778), Expect(2) = e-103
Identities = 134/173 (77%), Positives = 156/173 (89%)
Frame = -3
Query: 670 LGFERVLTDTDFSNLPYLQCVARNAMRLHPPTPLMLPHRSNANVKIGGYDIPKGSNVHVN 491
+G +RV+ +TDF +LPYLQC+ + A+RLHPPTPLMLPH++ NVKIGGYDIPKGSNVHVN
Sbjct: 338 VGRDRVMNETDFPHLPYLQCITKEALRLHPPTPLMLPHKATQNVKIGGYDIPKGSNVHVN 397
Query: 490 VWAVARDPAVWKNPLEFRPERFLEEDVDMKGHDFRLLPFGAGRRVCPGAQLGINLVTSML 311
VWA+ARDPAVWK+P+ FRPERFLEEDVD+KGHD+RLLPFGAGRR+CPGAQLGINLV SML
Sbjct: 398 VWAIARDPAVWKDPVTFRPERFLEEDVDIKGHDYRLLPFGAGRRICPGAQLGINLVQSML 457
Query: 310 GHLLHHFCWAPPEGVKPEEIDMVENPGLVTYMRTPVQAVATPRLPSHLYKRVP 152
GHLLHHF WAPPEG++ E+ID+ ENPGLVT+M PVQA+A PRLP HLYKR P
Sbjct: 458 GHLLHHFVWAPPEGMQAEDIDLTENPGLVTFMAKPVQAIAIPRLPDHLYKRQP 510
Score = 95.5 bits (236), Expect(2) = e-103
Identities = 44/52 (84%), Positives = 50/52 (95%)
Frame = -2
Query: 827 QYDLSEDTIIGLLWDMITAGMDTTAISVEWAMAELIKNPGLQPKAQEELDRV 672
+YDLSEDTIIGLLWDMITAGMDTTAI+VEWAMAEL++NP +Q KAQEE+DRV
Sbjct: 286 KYDLSEDTIIGLLWDMITAGMDTTAITVEWAMAELVRNPRIQQKAQEEIDRV 337
>emb|CAD20576.1| putative cytochrome P450 [Solenostemon scutellarioides]
Length = 507
Score = 300 bits (767), Expect(2) = e-100
Identities = 136/172 (79%), Positives = 154/172 (89%)
Frame = -3
Query: 670 LGFERVLTDTDFSNLPYLQCVARNAMRLHPPTPLMLPHRSNANVKIGGYDIPKGSNVHVN 491
+G ER++T+ D NLPYLQCV + ++RLHPPTP+MLPHRSNA+VKIGGYDIPKGSNVHVN
Sbjct: 331 IGHERIMTELDIPNLPYLQCVVKESLRLHPPTPVMLPHRSNADVKIGGYDIPKGSNVHVN 390
Query: 490 VWAVARDPAVWKNPLEFRPERFLEEDVDMKGHDFRLLPFGAGRRVCPGAQLGINLVTSML 311
VWA+ARDP WK PLEFRPERFLEEDVD+KGHDFRLL FGAGRRVCPGAQLGI+L TSM+
Sbjct: 391 VWAIARDPKSWKVPLEFRPERFLEEDVDIKGHDFRLLRFGAGRRVCPGAQLGIDLATSMI 450
Query: 310 GHLLHHFCWAPPEGVKPEEIDMVENPGLVTYMRTPVQAVATPRLPSHLYKRV 155
GHLLHHF W PP GV+ E+IDM ENPG VTYMRTPV+AV TPRLP++LYKRV
Sbjct: 451 GHLLHHFRWTPPTGVRAEDIDMGENPGTVTYMRTPVEAVPTPRLPANLYKRV 502
Score = 89.4 bits (220), Expect(2) = e-100
Identities = 42/52 (80%), Positives = 46/52 (87%)
Frame = -2
Query: 827 QYDLSEDTIIGLLWDMITAGMDTTAISVEWAMAELIKNPGLQPKAQEELDRV 672
+YDLSEDTII LLWDMI AGMDT AISVEWAMAEL++NP +Q K QEELDRV
Sbjct: 279 EYDLSEDTIIALLWDMIAAGMDTPAISVEWAMAELVRNPRVQEKVQEELDRV 330
>dbj|BAC44836.1| cytochrome P-450 [Lithospermum erythrorhizon]
Length = 506
Score = 285 bits (730), Expect(2) = 1e-96
Identities = 129/177 (72%), Positives = 153/177 (85%), Gaps = 1/177 (0%)
Frame = -3
Query: 670 LGFERVLTDTDFSNLPYLQCVARNAMRLHPPTPLMLPHRSNANVKIGGYDIPKGSNVHVN 491
+G +R++T+ D LPYLQC+ + ++RLHPPTPLMLPHR++ANVKIGGYDIPKGS VHVN
Sbjct: 330 VGPDRIMTEADVPKLPYLQCIVKESLRLHPPTPLMLPHRASANVKIGGYDIPKGSIVHVN 389
Query: 490 VWAVARDPAVWKNPLEFRPERFLEEDVDMKGHDFRLLPFGAGRRVCPGAQLGINLVTSML 311
VWA+ARDPA WKNP EFRPERF+EED+DMKG D+RLLPFGAGRR+CPGAQL INL+TS L
Sbjct: 390 VWAIARDPAYWKNPEEFRPERFMEEDIDMKGTDYRLLPFGAGRRICPGAQLAINLITSSL 449
Query: 310 GHLLHHFCWAPPEGVKPEEIDMVENPGLVTYMRTPVQAVATPRLPS-HLYKRVPADI 143
GHLLH F W+P GVKPEEID+ ENPG VTYMR PV+AV +PRL + HLYKRV +D+
Sbjct: 450 GHLLHQFTWSPQPGVKPEEIDLSENPGTVTYMRNPVKAVVSPRLSAVHLYKRVESDM 506
Score = 90.5 bits (223), Expect(2) = 1e-96
Identities = 42/52 (80%), Positives = 46/52 (87%)
Frame = -2
Query: 827 QYDLSEDTIIGLLWDMITAGMDTTAISVEWAMAELIKNPGLQPKAQEELDRV 672
QYD+SEDTIIGLLWDMI AGMDT IS EWAMAEL++NP +Q KAQEELDRV
Sbjct: 278 QYDISEDTIIGLLWDMIAAGMDTATISTEWAMAELVRNPRVQRKAQEELDRV 329
>sp|O48956|C981_SORBI Cytochrome P450 98A1 gi|7430624|pir||T14638 cytochrome P450 CYP98A1
- sorghum gi|2766448|gb|AAC39316.1| cytochrome P450
CYP98A1 [Sorghum bicolor]
Length = 512
Score = 278 bits (712), Expect(2) = 1e-95
Identities = 126/177 (71%), Positives = 154/177 (86%), Gaps = 1/177 (0%)
Frame = -3
Query: 670 LGFERVLTDTDFSNLPYLQCVARNAMRLHPPTPLMLPHRSNANVKIGGYDIPKGSNVHVN 491
+G +RV+ +TDF NLPYLQ V + ++RLHPPTPLMLPH+++ NVKIGGYDIPKG+NV VN
Sbjct: 336 VGRDRVMLETDFQNLPYLQAVVKESLRLHPPTPLMLPHKASTNVKIGGYDIPKGANVMVN 395
Query: 490 VWAVARDPAVWKNPLEFRPERFLEEDVDMKGHDFRLLPFGAGRRVCPGAQLGINLVTSML 311
VWAVARDP VW NPLE+RPERFLEE++D+KG DFR+LPFGAGRRVCPGAQLGINLV SM+
Sbjct: 396 VWAVARDPKVWSNPLEYRPERFLEENIDIKGSDFRVLPFGAGRRVCPGAQLGINLVASMI 455
Query: 310 GHLLHHFCWAPPEGVKPEEIDMVENPGLVTYMRTPVQAVATPRL-PSHLYKRVPADI 143
GHLLHHF W+ PEG +PE+++M+E+PGLVT+M TP+QAVA PRL LY RVP ++
Sbjct: 456 GHLLHHFEWSLPEGTRPEDVNMMESPGLVTFMGTPLQAVAKPRLEKEELYNRVPVEM 512
Score = 94.7 bits (234), Expect(2) = 1e-95
Identities = 44/52 (84%), Positives = 48/52 (91%)
Frame = -2
Query: 827 QYDLSEDTIIGLLWDMITAGMDTTAISVEWAMAELIKNPGLQPKAQEELDRV 672
QYDLSEDT+IGLLWDMITAGMDTT ISVEWAMAEL++NP +Q K QEELDRV
Sbjct: 284 QYDLSEDTVIGLLWDMITAGMDTTVISVEWAMAELVRNPRVQKKLQEELDRV 335
>sp|O48922|C982_SOYBN Cytochrome P450 98A2 gi|7430683|pir||T05937 cytochrome P450
monooxygenase 98A2p - soybean gi|2738998|gb|AAB94587.1|
CYP98A2p [Glycine max]
Length = 509
Score = 343 bits (881), Expect = 1e-93
Identities = 157/193 (81%), Positives = 173/193 (89%)
Frame = -3
Query: 721 LKTQACSQKLKRS*TG*LGFERVLTDTDFSNLPYLQCVARNAMRLHPPTPLMLPHRSNAN 542
++ QK++ +G ERV+T+ DFSNLPYLQCV + AMRLHPPTPLMLPHR+NAN
Sbjct: 317 IRNPRVQQKVQEELDRVIGLERVMTEADFSNLPYLQCVTKEAMRLHPPTPLMLPHRANAN 376
Query: 541 VKIGGYDIPKGSNVHVNVWAVARDPAVWKNPLEFRPERFLEEDVDMKGHDFRLLPFGAGR 362
VK+GGYDIPKGSNVHVNVWAVARDPAVWK+PLEFRPERFLEEDVDMKGHDFRLLPFG+GR
Sbjct: 377 VKVGGYDIPKGSNVHVNVWAVARDPAVWKDPLEFRPERFLEEDVDMKGHDFRLLPFGSGR 436
Query: 361 RVCPGAQLGINLVTSMLGHLLHHFCWAPPEGVKPEEIDMVENPGLVTYMRTPVQAVATPR 182
RVCPGAQLGINL SMLGHLLHHFCW PPEG+KPEEIDM ENPGLVTYMRTP+QAV +PR
Sbjct: 437 RVCPGAQLGINLAASMLGHLLHHFCWTPPEGMKPEEIDMGENPGLVTYMRTPIQAVVSPR 496
Query: 181 LPSHLYKRVPADI 143
LPSHLYKRVPA+I
Sbjct: 497 LPSHLYKRVPAEI 509
Score = 95.9 bits (237), Expect = 6e-19
Identities = 46/52 (88%), Positives = 49/52 (93%)
Frame = -2
Query: 827 QYDLSEDTIIGLLWDMITAGMDTTAISVEWAMAELIKNPGLQPKAQEELDRV 672
+YDLSEDTIIGLLWDMITAGMDTTAISVEWAMAELI+NP +Q K QEELDRV
Sbjct: 282 KYDLSEDTIIGLLWDMITAGMDTTAISVEWAMAELIRNPRVQQKVQEELDRV 333
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 798,080,494
Number of Sequences: 1393205
Number of extensions: 19828670
Number of successful extensions: 57926
Number of sequences better than 10.0: 3011
Number of HSP's better than 10.0 without gapping: 50217
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 55280
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 42643890261
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)