Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003071A_C01 KMC003071A_c01
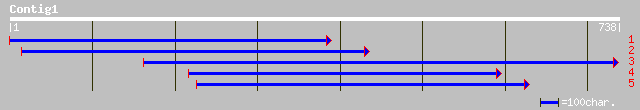
(735 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC49774.1| AP2 domain containing protein RAP2.8 [Arabidopsis... 138 7e-32
ref|NP_564947.1| AP2 domain protein RAP2.8 (RAV2); protein id: A... 138 7e-32
gb|AAM63120.1| putative RAV2-like DNA-binding protein [Arabidops... 138 9e-32
ref|NP_173927.1| DNA-binding protein RAV2, putative; protein id:... 135 5e-31
dbj|BAB84620.1| DNA-binding protein RAV2-like [Oryza sativa (jap... 132 7e-30
>gb|AAC49774.1| AP2 domain containing protein RAP2.8 [Arabidopsis thaliana]
Length = 334
Score = 138 bits (348), Expect = 7e-32
Identities = 82/147 (55%), Positives = 92/147 (61%), Gaps = 8/147 (5%)
Frame = -1
Query: 702 AAAKGMLLNFEDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDAVRFHRSTGP 523
A KG+L+NFEDV GKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGD V F RSTG
Sbjct: 204 AVTKGVLINFEDVNGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDVVTFERSTGL 263
Query: 522 DRQLYIDCKASSSSGGGVGEVVDVSIKTGGLFIPVGPVVEPVQ-MVRLFGV---NLLKLP 355
+RQLYID K S GP PVQ +VRLFGV N+ +
Sbjct: 264 ERQLYIDWKVRS-----------------------GPRENPVQVVVRLFGVDIFNVTTVK 300
Query: 354 GSD--GGCHGKRKE--MDLFSLECSQK 286
+D C GKR D+F+L CS+K
Sbjct: 301 PNDVVAVCGGKRSRDVDDMFALRCSKK 327
>ref|NP_564947.1| AP2 domain protein RAP2.8 (RAV2); protein id: At1g68840.1,
supported by cDNA: 19561., supported by cDNA:
gi_13430799, supported by cDNA: gi_15810644, supported
by cDNA: gi_3868858 [Arabidopsis thaliana]
gi|11357264|pir||T51330 DNA binding protein RAV2
[imported] - Arabidopsis thaliana
gi|3868859|dbj|BAA34251.1| RAV2 [Arabidopsis thaliana]
gi|12323214|gb|AAG51586.1|AC011665_7 putative
DNA-binding protein (RAV2-like) [Arabidopsis thaliana]
gi|12324134|gb|AAG52035.1|AC011914_5 RAV2; 17047-15989
[Arabidopsis thaliana]
gi|13430800|gb|AAK26022.1|AF360312_1 putative
DNA-binding protein(RAV2 [Arabidopsis thaliana]
gi|15810645|gb|AAL07247.1| putative DNA-binding protein
RAV2 [Arabidopsis thaliana]
Length = 352
Score = 138 bits (348), Expect = 7e-32
Identities = 82/147 (55%), Positives = 92/147 (61%), Gaps = 8/147 (5%)
Frame = -1
Query: 702 AAAKGMLLNFEDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDAVRFHRSTGP 523
A KG+L+NFEDV GKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGD V F RSTG
Sbjct: 222 AVTKGVLINFEDVNGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDVVTFERSTGL 281
Query: 522 DRQLYIDCKASSSSGGGVGEVVDVSIKTGGLFIPVGPVVEPVQ-MVRLFGV---NLLKLP 355
+RQLYID K S GP PVQ +VRLFGV N+ +
Sbjct: 282 ERQLYIDWKVRS-----------------------GPRENPVQVVVRLFGVDIFNVTTVK 318
Query: 354 GSD--GGCHGKRKE--MDLFSLECSQK 286
+D C GKR D+F+L CS+K
Sbjct: 319 PNDVVAVCGGKRSRDVDDMFALRCSKK 345
>gb|AAM63120.1| putative RAV2-like DNA-binding protein [Arabidopsis thaliana]
Length = 352
Score = 138 bits (347), Expect = 9e-32
Identities = 82/147 (55%), Positives = 91/147 (61%), Gaps = 8/147 (5%)
Frame = -1
Query: 702 AAAKGMLLNFEDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDAVRFHRSTGP 523
A KG+L+NFEDV GKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGD V F RSTG
Sbjct: 222 AVTKGVLINFEDVNGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDVVTFERSTGL 281
Query: 522 DRQLYIDCKASSSSGGGVGEVVDVSIKTGGLFIPVGPVVEPVQ-MVRLFGV---NLLKLP 355
+RQLYID K S GP PVQ +VRLFGV N+ +
Sbjct: 282 ERQLYIDWKVRS-----------------------GPRENPVQVVVRLFGVDIFNVTTVK 318
Query: 354 GSD--GGCHGKRKE--MDLFSLECSQK 286
+D C GKR D+F+L CS K
Sbjct: 319 PNDVVAVCGGKRSRDVDDMFALRCSNK 345
>ref|NP_173927.1| DNA-binding protein RAV2, putative; protein id: At1g25560.1,
supported by cDNA: gi_15810260, supported by cDNA:
gi_20259538 [Arabidopsis thaliana]
gi|25403024|pir||A86386 probable DNA-binding protein
RAV2 [imported] - Arabidopsis thaliana
gi|12321505|gb|AAG50808.1|AC079281_10 DNA-binding
protein RAV2, putative [Arabidopsis thaliana]
gi|20259539|gb|AAM13889.1| putative DNA-binding protein
RAV2 [Arabidopsis thaliana] gi|21689705|gb|AAM67474.1|
putative DNA-binding protein RAV2 [Arabidopsis thaliana]
Length = 361
Score = 135 bits (341), Expect = 5e-31
Identities = 80/144 (55%), Positives = 88/144 (60%), Gaps = 8/144 (5%)
Frame = -1
Query: 693 KGMLLNFEDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDAVRFHRSTGPDRQ 514
KG+L+N ED GKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGD V F RSTGPDRQ
Sbjct: 238 KGVLINLEDRTGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDVVCFERSTGPDRQ 297
Query: 513 LYIDCKASSSSGGGVGEVVDVSIKTGGLFIPVGPVVEPVQ-MVRLFGVNLLKLPGSDGG- 340
LYI K SS PVQ +VRLFGVN+ +
Sbjct: 298 LYIHWKVRSS---------------------------PVQTVVRLFGVNIFNVSNEKPND 330
Query: 339 ----CHGKR--KEMDLFSLECSQK 286
C GK+ +E DLFSL CS+K
Sbjct: 331 VAVECVGKKRSREDDLFSLGCSKK 354
>dbj|BAB84620.1| DNA-binding protein RAV2-like [Oryza sativa (japonica
cultivar-group)]
Length = 393
Score = 132 bits (331), Expect = 7e-30
Identities = 72/136 (52%), Positives = 83/136 (60%), Gaps = 3/136 (2%)
Frame = -1
Query: 708 ATAAAKGMLLNFEDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDAVRFHR-- 535
A +KG+LLNFED GKVWRFRYSYWNSSQSYVLTKGWSRFVKEK L AGD V F+R
Sbjct: 249 AGGESKGVLLNFEDAAGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKGLHAGDVVGFYRSA 308
Query: 534 -STGPDRQLYIDCKASSSSGGGVGEVVDVSIKTGGLFIPVGPVVEPVQMVRLFGVNLLKL 358
S G D +L+IDCK S+G + D P PV+ VRLFGV+LL
Sbjct: 309 ASAGDDGKLFIDCKLVRSTGAALASPAD------------QPAPSPVKAVRLFGVDLLTA 356
Query: 357 PGSDGGCHGKRKEMDL 310
P G ++ DL
Sbjct: 357 PAPVEQMAGCKRARDL 372
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 710,019,239
Number of Sequences: 1393205
Number of extensions: 17276508
Number of successful extensions: 71063
Number of sequences better than 10.0: 194
Number of HSP's better than 10.0 without gapping: 56765
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 67211
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 34906576228
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)