Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003067A_C01 KMC003067A_c01
(693 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
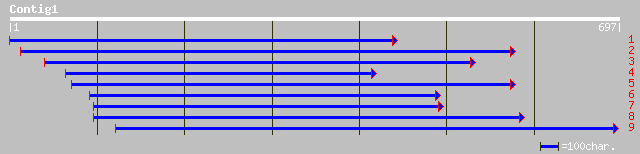
gb|AAK58876.1|AF307152_1 diphosphonucleotide phosphatase 2 [Zea ... 122 5e-54
dbj|BAA97052.1| gene_id:K15M2.3~unknown protein [Arabidopsis tha... 123 1e-52
gb|AAO41914.1| putative DNA nick sensor protein [Arabidopsis tha... 123 1e-52
gb|AAM69280.1|AF453835_1 phosphoesterase [Arabidopsis thaliana] 123 1e-52
ref|NP_188107.1| DNA nick sensor, putative; protein id: At3g1489... 123 9e-50
>gb|AAK58876.1|AF307152_1 diphosphonucleotide phosphatase 2 [Zea mays]
Length = 232
Score = 122 bits (307), Expect(2) = 5e-54
Identities = 60/80 (75%), Positives = 66/80 (82%)
Frame = -3
Query: 691 PDKLQSLYNNGYKLVIFTXESNIERWKKSRQKAVDSKIGRLNNFIEKVKVPIQVFIACGL 512
P+KLQ LY++GYKLVIFT ESNIERWK RQ+AVDSK+GRL+NFIE VK PIQVFIACGL
Sbjct: 99 PEKLQRLYDDGYKLVIFTNESNIERWKNKRQQAVDSKVGRLDNFIECVKAPIQVFIACGL 158
Query: 511 SNSGKAAGKEDDPYRKPKPG 452
GK G DDPYRKP PG
Sbjct: 159 ---GKGKGIPDDPYRKPNPG 175
Score = 110 bits (276), Expect(2) = 5e-54
Identities = 50/59 (84%), Positives = 55/59 (92%)
Frame = -1
Query: 462 PNQGMWQLMEQHFNSGITIDMDQSFYVGDAAGRKSDHSDADIKFAEAIGLKFHLPEEYF 286
PN GMW LM QHFNSGI IDMDQSFYVGDAAGR++DHSDADI+FA+AIGLKFH+PEEYF
Sbjct: 172 PNPGMWWLMAQHFNSGIKIDMDQSFYVGDAAGRENDHSDADIEFAKAIGLKFHVPEEYF 230
>dbj|BAA97052.1| gene_id:K15M2.3~unknown protein [Arabidopsis thaliana]
Length = 774
Score = 123 bits (308), Expect(2) = 1e-52
Identities = 61/80 (76%), Positives = 69/80 (86%)
Frame = -3
Query: 691 PDKLQSLYNNGYKLVIFTXESNIERWKKSRQKAVDSKIGRLNNFIEKVKVPIQVFIACGL 512
P+KLQSL++ GYKLVIFT ESNI+RWK RQ AVDSKIGRLN+FIE+VKVPIQVFIACG+
Sbjct: 632 PEKLQSLHDQGYKLVIFTNESNIDRWKNKRQAAVDSKIGRLNSFIERVKVPIQVFIACGV 691
Query: 511 SNSGKAAGKEDDPYRKPKPG 452
S+SG GK DD YRKPK G
Sbjct: 692 SSSGGKGGK-DDLYRKPKAG 710
Score = 106 bits (264), Expect(2) = 1e-52
Identities = 49/63 (77%), Positives = 53/63 (83%)
Frame = -1
Query: 474 LIANPNQGMWQLMEQHFNSGITIDMDQSFYVGDAAGRKSDHSDADIKFAEAIGLKFHLPE 295
L P GMWQLM++HFNSGI IDMD+SFYVGDAAGRK DHSDADIKFA+A GLKF PE
Sbjct: 703 LYRKPKAGMWQLMKKHFNSGIAIDMDKSFYVGDAAGRKMDHSDADIKFAQASGLKFFTPE 762
Query: 294 EYF 286
EYF
Sbjct: 763 EYF 765
>gb|AAO41914.1| putative DNA nick sensor protein [Arabidopsis thaliana]
gi|28827696|gb|AAO50692.1| putative DNA nick sensor
protein [Arabidopsis thaliana]
Length = 694
Score = 123 bits (308), Expect(2) = 1e-52
Identities = 61/80 (76%), Positives = 69/80 (86%)
Frame = -3
Query: 691 PDKLQSLYNNGYKLVIFTXESNIERWKKSRQKAVDSKIGRLNNFIEKVKVPIQVFIACGL 512
P+KLQSL++ GYKLVIFT ESNI+RWK RQ AVDSKIGRLN+FIE+VKVPIQVFIACG+
Sbjct: 552 PEKLQSLHDQGYKLVIFTNESNIDRWKNKRQAAVDSKIGRLNSFIERVKVPIQVFIACGV 611
Query: 511 SNSGKAAGKEDDPYRKPKPG 452
S+SG GK DD YRKPK G
Sbjct: 612 SSSGGKGGK-DDLYRKPKAG 630
Score = 106 bits (264), Expect(2) = 1e-52
Identities = 49/63 (77%), Positives = 53/63 (83%)
Frame = -1
Query: 474 LIANPNQGMWQLMEQHFNSGITIDMDQSFYVGDAAGRKSDHSDADIKFAEAIGLKFHLPE 295
L P GMWQLM++HFNSGI IDMD+SFYVGDAAGRK DHSDADIKFA+A GLKF PE
Sbjct: 623 LYRKPKAGMWQLMKKHFNSGIAIDMDKSFYVGDAAGRKMDHSDADIKFAQASGLKFFTPE 682
Query: 294 EYF 286
EYF
Sbjct: 683 EYF 685
>gb|AAM69280.1|AF453835_1 phosphoesterase [Arabidopsis thaliana]
Length = 638
Score = 123 bits (308), Expect(2) = 1e-52
Identities = 61/80 (76%), Positives = 69/80 (86%)
Frame = -3
Query: 691 PDKLQSLYNNGYKLVIFTXESNIERWKKSRQKAVDSKIGRLNNFIEKVKVPIQVFIACGL 512
P+KLQSL++ GYKLVIFT ESNI+RWK RQ AVDSKIGRLN+FIE+VKVPIQVFIACG+
Sbjct: 496 PEKLQSLHDQGYKLVIFTNESNIDRWKNKRQAAVDSKIGRLNSFIERVKVPIQVFIACGV 555
Query: 511 SNSGKAAGKEDDPYRKPKPG 452
S+SG GK DD YRKPK G
Sbjct: 556 SSSGGKGGK-DDLYRKPKAG 574
Score = 106 bits (264), Expect(2) = 1e-52
Identities = 49/63 (77%), Positives = 53/63 (83%)
Frame = -1
Query: 474 LIANPNQGMWQLMEQHFNSGITIDMDQSFYVGDAAGRKSDHSDADIKFAEAIGLKFHLPE 295
L P GMWQLM++HFNSGI IDMD+SFYVGDAAGRK DHSDADIKFA+A GLKF PE
Sbjct: 567 LYRKPKAGMWQLMKKHFNSGIAIDMDKSFYVGDAAGRKMDHSDADIKFAQASGLKFFTPE 626
Query: 294 EYF 286
EYF
Sbjct: 627 EYF 629
>ref|NP_188107.1| DNA nick sensor, putative; protein id: At3g14890.1 [Arabidopsis
thaliana]
Length = 640
Score = 123 bits (308), Expect(2) = 9e-50
Identities = 61/80 (76%), Positives = 69/80 (86%)
Frame = -3
Query: 691 PDKLQSLYNNGYKLVIFTXESNIERWKKSRQKAVDSKIGRLNNFIEKVKVPIQVFIACGL 512
P+KLQSL++ GYKLVIFT ESNI+RWK RQ AVDSKIGRLN+FIE+VKVPIQVFIACG+
Sbjct: 483 PEKLQSLHDQGYKLVIFTNESNIDRWKNKRQAAVDSKIGRLNSFIERVKVPIQVFIACGV 542
Query: 511 SNSGKAAGKEDDPYRKPKPG 452
S+SG GK DD YRKPK G
Sbjct: 543 SSSGGKGGK-DDLYRKPKAG 561
Score = 96.3 bits (238), Expect(2) = 9e-50
Identities = 49/78 (62%), Positives = 53/78 (67%), Gaps = 15/78 (19%)
Frame = -1
Query: 474 LIANPNQGMWQLMEQHFNSGITIDMD---------------QSFYVGDAAGRKSDHSDAD 340
L P GMWQLM++HFNSGI IDMD +SFYVGDAAGRK DHSDAD
Sbjct: 554 LYRKPKAGMWQLMKKHFNSGIAIDMDNTFFVVDTDICAMLIRSFYVGDAAGRKMDHSDAD 613
Query: 339 IKFAEAIGLKFHLPEEYF 286
IKFA+A GLKF PEEYF
Sbjct: 614 IKFAQASGLKFFTPEEYF 631
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 555,678,465
Number of Sequences: 1393205
Number of extensions: 11497453
Number of successful extensions: 25149
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 24429
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25120
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31401661572
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)