Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
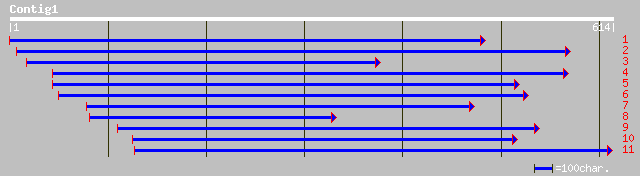
Query= KMC003065A_C01 KMC003065A_c01
(611 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T09704 probable arginine/serine-rich splicing factor - alfa... 97 2e-19
dbj|BAB64646.1| putative splicing factor-like protein [Oryza sat... 56 4e-07
pir||S50755 hypothetical protein VSP-3 - Chlamydomonas reinhardt... 55 9e-07
ref|NP_068239.1| PxORF20 peptide [Plutella xylostella granulovir... 54 2e-06
ref|NP_495013.1| pre-mRNA splicing SR Protein RSP-4 (22.6 kD) (r... 52 8e-06
>pir||T09704 probable arginine/serine-rich splicing factor - alfalfa
gi|3334756|emb|CAA76346.1| putative arginine/serine-rich
splicing factor [Medicago sativa]
Length = 286
Score = 96.7 bits (239), Expect = 2e-19
Identities = 61/101 (60%), Positives = 62/101 (60%), Gaps = 24/101 (23%)
Frame = -3
Query: 609 SVDSRSPAR------RSPSPRRSPSPKRSVSPKR------------------STSPRKSP 502
SVDSRSP R RSPSPRRSPSPKRS SPKR S SPRKSP
Sbjct: 192 SVDSRSPVRRSSIPKRSPSPRRSPSPKRSPSPKRSPSPKRSPSLKRSISPQKSVSPRKSP 251
Query: 501 RGESPDNRSRDGRSPTPRSVSPRGRPVASRSPSPRKSNGDE 379
ESPD+RSR GRS TPRS P ASRSPSPR SNGDE
Sbjct: 252 LRESPDSRSRGGRSFTPRS------PGASRSPSPRNSNGDE 286
Score = 37.4 bits (85), Expect = 0.16
Identities = 26/69 (37%), Positives = 31/69 (44%)
Frame = -3
Query: 597 RSPARRSPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSVSPRGRPVA 418
R RRS S SP K + R R +RS D RSP RS P+ P
Sbjct: 157 RDHRRRSRSRSASPGYKGRGRGRHDDERRSRSR-----SRSVDSRSPVRRSSIPKRSPSP 211
Query: 417 SRSPSPRKS 391
RSPSP++S
Sbjct: 212 RRSPSPKRS 220
>dbj|BAB64646.1| putative splicing factor-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 306
Score = 55.8 bits (133), Expect = 4e-07
Identities = 38/90 (42%), Positives = 46/90 (50%), Gaps = 20/90 (22%)
Frame = -3
Query: 600 SRSPARRSPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRS---------------RDG 466
S+SP R S + RRS SP+ S SP+ S +SPRG D+RS RD
Sbjct: 168 SQSPRRDSRNERRSRSPRDSRSPRGSPRDSRSPRGSPRDSRSPKGSPRDTQSPRGSPRDS 227
Query: 465 RSPTPRSVSPRGR-----PVASRSPSPRKS 391
RSP + P GR P ASRSP+PR S
Sbjct: 228 RSPRRSASPPNGRNRSPTPNASRSPAPRDS 257
Score = 52.8 bits (125), Expect = 4e-06
Identities = 42/91 (46%), Positives = 49/91 (53%), Gaps = 6/91 (6%)
Frame = -3
Query: 600 SRSPA-RRSPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSP--TPR-SVSPR 433
SRSP+ RR S RS S RS S RS SPR+ R E RD RSP +PR S SPR
Sbjct: 142 SRSPSPRRGRSRGRSYSRSRSRSYSRSQSPRRDSRNERRSRSPRDSRSPRGSPRDSRSPR 201
Query: 432 GRPVASRSP--SPRKSNGDE*TVFNEKFQRR 346
G P SRSP SPR + + + + RR
Sbjct: 202 GSPRDSRSPKGSPRDTQSPRGSPRDSRSPRR 232
Score = 42.0 bits (97), Expect = 0.006
Identities = 28/74 (37%), Positives = 37/74 (49%), Gaps = 11/74 (14%)
Frame = -3
Query: 603 DSRSPAR---------RSPSPR--RSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSP 457
DSRSP R RSP+P RSP+P+ S SP R+ S + + + +GRSP
Sbjct: 226 DSRSPRRSASPPNGRNRSPTPNASRSPAPRDSRSPMRADSRSPADHERRDMSTAANGRSP 285
Query: 456 TPRSVSPRGRPVAS 415
+PR G AS
Sbjct: 286 SPRDYEDNGNHRAS 299
Score = 42.0 bits (97), Expect = 0.006
Identities = 36/90 (40%), Positives = 42/90 (46%), Gaps = 16/90 (17%)
Frame = -3
Query: 603 DSRSPA---RRSPSPRRSP----SPKRSVSP----KRSTSPRKSPRGESPDNRS---RDG 466
DSRSP R + SPR SP SP+RS SP RS +P S D+RS D
Sbjct: 206 DSRSPKGSPRDTQSPRGSPRDSRSPRRSASPPNGRNRSPTPNASRSPAPRDSRSPMRADS 265
Query: 465 RSPTPRSVSPRGRPVASRSPSPR--KSNGD 382
RSP RSPSPR + NG+
Sbjct: 266 RSPADHERRDMSTAANGRSPSPRDYEDNGN 295
>pir||S50755 hypothetical protein VSP-3 - Chlamydomonas reinhardtii
gi|530876|gb|AAB53953.1| amino acid feature: Rod protein
domain, aa 266 .. 468; amino acid feature: globular
protein domain, aa 32 .. 265
Length = 473
Score = 54.7 bits (130), Expect = 9e-07
Identities = 32/73 (43%), Positives = 42/73 (56%), Gaps = 3/73 (4%)
Frame = -3
Query: 600 SRSPARRSPSPRRSPSPKRSVSPKRSTSPRKSPRGE---SPDNRSRDGRSPTPRSVSPRG 430
+R+ A SPSP+ SPSPK S SP SP SP+ SP ++ SP+P++ SP
Sbjct: 261 NRTGASPSPSPKASPSPKVSPSPSPKASPSPSPKASPSPSPSPKASPSPSPSPKA-SPSP 319
Query: 429 RPVASRSPSPRKS 391
P S SPSP+ S
Sbjct: 320 SPSPSPSPSPKAS 332
Score = 50.8 bits (120), Expect = 1e-05
Identities = 31/63 (49%), Positives = 35/63 (55%)
Frame = -3
Query: 579 SPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSVSPRGRPVASRSPSP 400
SPSP+ SPSP SP S SP+ SP SP + SP+P S SP P AS SPSP
Sbjct: 282 SPSPKASPSPSPKASPSPSPSPKASP---SPSPSPKASPSPSP-SPSPSPSPKASPSPSP 337
Query: 399 RKS 391
S
Sbjct: 338 SPS 340
Score = 50.8 bits (120), Expect = 1e-05
Identities = 32/63 (50%), Positives = 37/63 (57%)
Frame = -3
Query: 579 SPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSVSPRGRPVASRSPSP 400
SPSP+ SPSP S SPK S SP SP SP ++ SP+P SV P +P S SPSP
Sbjct: 300 SPSPKASPSP--SPSPKASPSPSPSP-SPSPSPKASPSPSPSP-SVQPASKPSPSPSPSP 355
Query: 399 RKS 391
S
Sbjct: 356 SPS 358
Score = 49.3 bits (116), Expect = 4e-05
Identities = 31/63 (49%), Positives = 34/63 (53%)
Frame = -3
Query: 579 SPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSVSPRGRPVASRSPSP 400
SPSP SPSPK S SP S SP P SP SP+P+ VSP P S SPSP
Sbjct: 375 SPSPSPSPSPKPSPSPSPSPSPSPKP---SPSPSPSPSPSPSPK-VSPSPSPSPSPSPSP 430
Query: 399 RKS 391
+ S
Sbjct: 431 KAS 433
Score = 48.9 bits (115), Expect = 5e-05
Identities = 32/69 (46%), Positives = 35/69 (50%), Gaps = 1/69 (1%)
Frame = -3
Query: 594 SPARRSPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPR-SVSPRGRPVA 418
SP SPSP SPSP S SP SP SP SP + SP+P S SP+ P
Sbjct: 362 SPPLPSPSPSPSPSPSPSPSPSPKPSPSPSP-SPSPSPKPSPSPSPSPSPSPSPKVSPSP 420
Query: 417 SRSPSPRKS 391
S SPSP S
Sbjct: 421 SPSPSPSPS 429
Score = 48.1 bits (113), Expect = 9e-05
Identities = 32/71 (45%), Positives = 35/71 (49%), Gaps = 1/71 (1%)
Frame = -3
Query: 600 SRSPARRSPSPRRSPSPKRSVSPKRSTSPRKSPR-GESPDNRSRDGRSPTPRSVSPRGRP 424
S SP P P SPSP S SP S SP+ SP SP + SP+P S SP P
Sbjct: 356 SPSPRPSPPLPSPSPSPSPSPSPSPSPSPKPSPSPSPSPSPSPKPSPSPSP-SPSPSPSP 414
Query: 423 VASRSPSPRKS 391
S SPSP S
Sbjct: 415 KVSPSPSPSPS 425
Score = 46.2 bits (108), Expect = 3e-04
Identities = 34/76 (44%), Positives = 38/76 (49%), Gaps = 3/76 (3%)
Frame = -3
Query: 609 SVDSRSPARRSPSPRRSPSPKRS---VSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSVS 439
SV S SPSP SPSP+ S SP S SP SP SP + SP+P S S
Sbjct: 340 SVQPASKPSPSPSPSPSPSPRPSPPLPSPSPSPSPSPSP-SPSPSPKPSPSPSPSP-SPS 397
Query: 438 PRGRPVASRSPSPRKS 391
P+ P S SPSP S
Sbjct: 398 PKPSPSPSPSPSPSPS 413
Score = 45.1 bits (105), Expect = 7e-04
Identities = 29/60 (48%), Positives = 34/60 (56%)
Frame = -3
Query: 579 SPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSVSPRGRPVASRSPSP 400
SPSP SPSPK S SP S SP SP+ SP SP+P S SP+ P ++ PSP
Sbjct: 389 SPSPSPSPSPKPSPSPSPSPSPSPSPK-VSPS----PSPSPSP-SPSPKASPSPAKKPSP 442
Score = 44.7 bits (104), Expect = 0.001
Identities = 33/74 (44%), Positives = 37/74 (49%), Gaps = 4/74 (5%)
Frame = -3
Query: 600 SRSPA-RRSPSPRRSPS--PKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPR-SVSPR 433
S SP+ + SPSP SPS P SP S SP SPR P SP+P S SP
Sbjct: 324 SPSPSPKASPSPSPSPSVQPASKPSPSPSPSPSPSPRPSPPLPSPSPSPSPSPSPSPSPS 383
Query: 432 GRPVASRSPSPRKS 391
+P S SPSP S
Sbjct: 384 PKPSPSPSPSPSPS 397
Score = 40.8 bits (94), Expect = 0.014
Identities = 24/60 (40%), Positives = 26/60 (43%)
Frame = -3
Query: 579 SPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSVSPRGRPVASRSPSP 400
SPSP SPSP VSP S SP SP SP + P+P G P P P
Sbjct: 403 SPSPSPSPSPSPKVSPSPSPSPSPSP---SPKASPSPAKKPSPPPPVEEGAPPPIEGPPP 459
Score = 35.0 bits (79), Expect = 0.77
Identities = 26/69 (37%), Positives = 30/69 (42%), Gaps = 4/69 (5%)
Frame = -3
Query: 579 SPSPRR----SPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSVSPRGRPVASR 412
SPSP+ SPSP S SPK S SP K P SP +G P P +
Sbjct: 411 SPSPKVSPSPSPSPSPSPSPKASPSPAKKP---SPPPPVEEGAPPPIEGPPPMEE---AA 464
Query: 411 SPSPRKSNG 385
P P+K G
Sbjct: 465 PPPPKKKTG 473
>ref|NP_068239.1| PxORF20 peptide [Plutella xylostella granulovirus]
gi|11036815|gb|AAG27318.1|AF270937_20 PxORF20 peptide
[Plutella xylostella granulovirus]
Length = 235
Score = 53.9 bits (128), Expect = 2e-06
Identities = 36/71 (50%), Positives = 41/71 (57%), Gaps = 7/71 (9%)
Frame = -3
Query: 585 RRSPSPRRSPSPKRSVSPKRSTSP-----RKSPRGESPDNRSRDGR--SPTPRSVSPRGR 427
RR PSPRR PSP+R SP+R SP R S R SP RS R SP+ R SPR R
Sbjct: 107 RRCPSPRRCPSPRRCPSPRRCPSPHVGRRRSSRRCPSPRRRSSTKRCQSPSKRCSSPRRR 166
Query: 426 PVASRSPSPRK 394
+SR SPR+
Sbjct: 167 --SSRCESPRR 175
Score = 46.6 bits (109), Expect = 3e-04
Identities = 32/87 (36%), Positives = 38/87 (42%), Gaps = 23/87 (26%)
Frame = -3
Query: 585 RRSPSPRRSPSPKRSVSPKRSTSPRKSP-----------------------RGESPDNRS 475
RR PSPRR PSP+R SP+R SPR+ P R +SP R
Sbjct: 101 RRCPSPRRCPSPRRCPSPRRCPSPRRCPSPHVGRRRSSRRCPSPRRRSSTKRCQSPSKRC 160
Query: 474 RDGRSPTPRSVSPRGRPVASRSPSPRK 394
R + R SPR RS SPR+
Sbjct: 161 SSPRRRSSRCESPR-----RRSSSPRR 182
Score = 43.1 bits (100), Expect = 0.003
Identities = 29/65 (44%), Positives = 34/65 (51%), Gaps = 12/65 (18%)
Frame = -3
Query: 585 RRSPSPRRSPSP--------KRSVSPKRSTSPRK----SPRGESPDNRSRDGRSPTPRSV 442
RR PSPRR PSP +R SP+R +S ++ S R SP RS SP RS
Sbjct: 119 RRCPSPRRCPSPHVGRRRSSRRCPSPRRRSSTKRCQSPSKRCSSPRRRSSRCESPRRRSS 178
Query: 441 SPRGR 427
SPR R
Sbjct: 179 SPRRR 183
Score = 41.6 bits (96), Expect = 0.008
Identities = 29/73 (39%), Positives = 33/73 (44%), Gaps = 9/73 (12%)
Frame = -3
Query: 585 RRSPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTP---------RSVSPR 433
RR S R PSP+R SP+R SPR+ P SP R P+P R SPR
Sbjct: 89 RRRRSAPRCPSPRRCPSPRRCPSPRRCP---SPRRCPSPRRCPSPHVGRRRSSRRCPSPR 145
Query: 432 GRPVASRSPSPRK 394
R R SP K
Sbjct: 146 RRSSTKRCQSPSK 158
Score = 36.6 bits (83), Expect = 0.27
Identities = 29/65 (44%), Positives = 34/65 (51%), Gaps = 3/65 (4%)
Frame = -3
Query: 579 SPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSV-SPR-GRPVAS-RS 409
SP RR S R SP+R SPR+ P SP R P+PR SP GR +S R
Sbjct: 85 SPHRRRRRSAPRCPSPRRCPSPRRCP---SPRRCPSPRRCPSPRRCPSPHVGRRRSSRRC 141
Query: 408 PSPRK 394
PSPR+
Sbjct: 142 PSPRR 146
Score = 33.9 bits (76), Expect = 1.7
Identities = 24/53 (45%), Positives = 28/53 (52%), Gaps = 8/53 (15%)
Frame = -3
Query: 606 VDSRSPARRSPSPRR--------SPSPKRSVSPKRSTSPRKSPRGESPDNRSR 472
V R +RR PSPRR SPS KR SP+R +S +SPR S R R
Sbjct: 132 VGRRRSSRRCPSPRRRSSTKRCQSPS-KRCSSPRRRSSRCESPRRRSSSPRRR 183
>ref|NP_495013.1| pre-mRNA splicing SR Protein RSP-4 (22.6 kD) (rsp-4)
[Caenorhabditis elegans] gi|3929375|sp|Q09511|SFR2_CAEEL
Putative splicing factor, arginine/serine-rich 2
(Splicing factor SC35) (SC-35) (Splicing component, 35
kDa) gi|7439997|pir||T15917 hypothetical protein EEED8.7
- Caenorhabditis elegans gi|733604|gb|AAC46767.1| Sr
protein (splicing factor) protein 4, isoform a
[Caenorhabditis elegans]
Length = 196
Score = 51.6 bits (122), Expect = 8e-06
Identities = 41/81 (50%), Positives = 46/81 (56%), Gaps = 10/81 (12%)
Frame = -3
Query: 600 SRSPARRSPSPR--RSPSPKRSVSPKRSTSPRKSPRGESPDNRSRD---GRSPTPR-SVS 439
SRSP RRS SPR RS SP+RS S R+ SP R +SPD R RSP PR S
Sbjct: 116 SRSPRRRSRSPRYSRSRSPRRSRS--RTRSPPSRDRRDSPDRRDNSRSRSRSPPPREDGS 173
Query: 438 PRGR----PVASRSPSPRKSN 388
P+ R ASRSPS +SN
Sbjct: 174 PKERRSRSRSASRSPSRSRSN 194
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 535,468,957
Number of Sequences: 1393205
Number of extensions: 12426765
Number of successful extensions: 77442
Number of sequences better than 10.0: 1629
Number of HSP's better than 10.0 without gapping: 49029
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 67476
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24568846532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)