Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003064A_C01 KMC003064A_c01
(1013 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
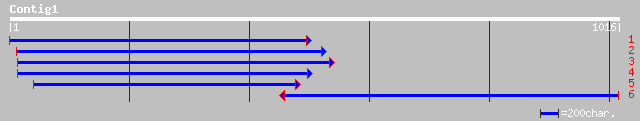
gb|AAM61357.1| unknown [Arabidopsis thaliana] 310 2e-83
ref|NP_565539.1| expressed protein; protein id: At2g22570.1, sup... 308 9e-83
gb|AAK84475.1| unknown [Lycopersicon esculentum] 291 8e-78
pir||C84614 hypothetical protein At2g22570 [imported] - Arabidop... 249 6e-65
ref|ZP_00073846.1| hypothetical protein [Trichodesmium erythraeu... 232 6e-60
>gb|AAM61357.1| unknown [Arabidopsis thaliana]
Length = 244
Score = 310 bits (795), Expect = 2e-83
Identities = 139/219 (63%), Positives = 179/219 (81%)
Frame = -1
Query: 947 LSGDIKTGLVLVDVVNGFCTVGAGNMAPTEPNEKITKMVEESVSLSKKFAEKNWPIFAFL 768
L+ D GLV+VDVVNGFCT+G+GNMAPT+ NE+I+KMVEES L+++F ++ WP+ AF+
Sbjct: 25 LNRDSSVGLVIVDVVNGFCTIGSGNMAPTKHNEQISKMVEESAKLAREFCDRKWPVLAFI 84
Query: 767 DSHHPDIPEPPYPSHCLIGSDEGKLVPELLWLENEPNATLRRKECIDGFLGSYEKDGSNV 588
DSHHPDIPE PYP HC+IG++E +LVP L WLE+E ATLRRK+CI+GF+GS E DGSNV
Sbjct: 85 DSHHPDIPERPYPPHCIIGTEESELVPALKWLESEDCATLRRKDCINGFVGSMESDGSNV 144
Query: 587 FADWVKKNQIKQILVAGICTDVCVLDFVCSVLSARNRQFLPPLENVIVSTEACSTYDVPL 408
F DWVK+NQIK I+V GICTD+CV DFV + LSARN L P+E+V+V + C+T+D+PL
Sbjct: 145 FVDWVKENQIKVIVVVGICTDICVFDFVATALSARNHGVLSPVEDVVVYSRGCATFDLPL 204
Query: 407 HVAKTNKDWVSHPQELMHHIGLYIACGRGAEIASEVIFE 291
HVAK K +HPQELMHH+GLY+A GRGA++ S++ FE
Sbjct: 205 HVAKDIKGAQAHPQELMHHVGLYMAKGRGAQVVSKISFE 243
>ref|NP_565539.1| expressed protein; protein id: At2g22570.1, supported by cDNA:
117955. [Arabidopsis thaliana]
gi|20197887|gb|AAM15300.1| expressed protein
[Arabidopsis thaliana] gi|26983880|gb|AAN86192.1|
unknown protein [Arabidopsis thaliana]
Length = 244
Score = 308 bits (789), Expect = 9e-83
Identities = 138/219 (63%), Positives = 178/219 (81%)
Frame = -1
Query: 947 LSGDIKTGLVLVDVVNGFCTVGAGNMAPTEPNEKITKMVEESVSLSKKFAEKNWPIFAFL 768
L+ D GLV+VDVVNGFCT+G+GNMAPT+ NE+I+KMVEES L+++F ++ WP+ AF+
Sbjct: 25 LNRDSSVGLVIVDVVNGFCTIGSGNMAPTKHNEQISKMVEESAKLAREFCDRKWPVLAFI 84
Query: 767 DSHHPDIPEPPYPSHCLIGSDEGKLVPELLWLENEPNATLRRKECIDGFLGSYEKDGSNV 588
DSHHPDIPE PYP HC+IG++E +LVP L WLE+E ATLRRK+CI+GF+GS E DGSNV
Sbjct: 85 DSHHPDIPERPYPPHCIIGTEESELVPALKWLESEDCATLRRKDCINGFVGSMESDGSNV 144
Query: 587 FADWVKKNQIKQILVAGICTDVCVLDFVCSVLSARNRQFLPPLENVIVSTEACSTYDVPL 408
F DWVK+ QIK I+V GICTD+CV DFV + LSARN L P+E+V+V + C+T+D+PL
Sbjct: 145 FVDWVKEKQIKVIVVVGICTDICVFDFVATALSARNHGVLSPVEDVVVYSRGCATFDLPL 204
Query: 407 HVAKTNKDWVSHPQELMHHIGLYIACGRGAEIASEVIFE 291
HVAK K +HPQELMHH+GLY+A GRGA++ S++ FE
Sbjct: 205 HVAKDIKGAQAHPQELMHHVGLYMAKGRGAQVVSKISFE 243
>gb|AAK84475.1| unknown [Lycopersicon esculentum]
Length = 230
Score = 291 bits (746), Expect = 8e-78
Identities = 132/195 (67%), Positives = 160/195 (81%)
Frame = -1
Query: 947 LSGDIKTGLVLVDVVNGFCTVGAGNMAPTEPNEKITKMVEESVSLSKKFAEKNWPIFAFL 768
L+GD+ TGLVLVD+VNGFCTVGAGN+AP PN +I+ MV+ESV L+K F EK WPI+A
Sbjct: 27 LTGDVNTGLVLVDIVNGFCTVGAGNLAPVTPNRQISAMVDESVKLAKVFCEKKWPIYALR 86
Query: 767 DSHHPDIPEPPYPSHCLIGSDEGKLVPELLWLENEPNATLRRKECIDGFLGSYEKDGSNV 588
DSHHPD+PEPP P HC+ G+DE +LVP L WLENEPN T+R K+CIDGFLGS EKDGSNV
Sbjct: 87 DSHHPDVPEPPNPPHCIAGTDESELVPALQWLENEPNVTVRCKDCIDGFLGSIEKDGSNV 146
Query: 587 FADWVKKNQIKQILVAGICTDVCVLDFVCSVLSARNRQFLPPLENVIVSTEACSTYDVPL 408
F +WVK N+IK ILV GICTD+CVLDFVCSVLSARNR FL PL++VIV + C+TYD+P+
Sbjct: 147 FVNWVKANEIKIILVVGICTDICVLDFVCSVLSARNRGFLSPLKDVIVYSPGCATYDLPV 206
Query: 407 HVAKTNKDWVSHPQE 363
+A+ K + HPQ+
Sbjct: 207 QIARNIKGALPHPQD 221
>pir||C84614 hypothetical protein At2g22570 [imported] - Arabidopsis thaliana
gi|4314355|gb|AAD15566.1| unknown protein [Arabidopsis
thaliana]
Length = 226
Score = 249 bits (635), Expect = 6e-65
Identities = 115/194 (59%), Positives = 146/194 (74%)
Frame = -1
Query: 947 LSGDIKTGLVLVDVVNGFCTVGAGNMAPTEPNEKITKMVEESVSLSKKFAEKNWPIFAFL 768
L+ D GLV+VDVVNGFCT+G+GNM MVEES L+++F ++ WP+ AF+
Sbjct: 25 LNRDSSVGLVIVDVVNGFCTIGSGNM-----------MVEESAKLAREFCDRKWPVLAFI 73
Query: 767 DSHHPDIPEPPYPSHCLIGSDEGKLVPELLWLENEPNATLRRKECIDGFLGSYEKDGSNV 588
DSHHPDIPE PYP HC+IG++E +LVP L WLE+E ATLRRK+CI+GF+GS E DGSNV
Sbjct: 74 DSHHPDIPERPYPPHCIIGTEESELVPALKWLESEDCATLRRKDCINGFVGSMESDGSNV 133
Query: 587 FADWVKKNQIKQILVAGICTDVCVLDFVCSVLSARNRQFLPPLENVIVSTEACSTYDVPL 408
F DWVK+ QIK I+V GICTD+CV DFV + LSARN L P+E+V+V + C+T+D+PL
Sbjct: 134 FVDWVKEKQIKVIVVVGICTDICVFDFVATALSARNHGVLSPVEDVVVYSRGCATFDLPL 193
Query: 407 HVAKTNKDWVSHPQ 366
HVAK K +HPQ
Sbjct: 194 HVAKDIKGAQAHPQ 207
>ref|ZP_00073846.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 247
Score = 232 bits (592), Expect = 6e-60
Identities = 111/216 (51%), Positives = 148/216 (68%), Gaps = 3/216 (1%)
Frame = -1
Query: 938 DIKTGLVLVDVVNGFCTVGAGNMAPTEPNEKITKMVEESVSLSKKFAEKNWPIFAFLDSH 759
D TGL++VDV+NGFCTVG G +AP EPNE+I MV ES L++ F EK WP+ AFLD+H
Sbjct: 30 DRPTGLIVVDVLNGFCTVGFGPLAPQEPNEQIATMVSESDRLARTFVEKGWPVLAFLDTH 89
Query: 758 HPDIPEPPYPSHCLIGSDEGKLVPELLWLENEPNATLRRKECIDGFLGSYEKD-GSNVFA 582
P PEPPYP HC G+ E +LVPEL WL + P ATL K+CI+GF+GS + D NV
Sbjct: 90 EPGKPEPPYPPHCEKGTGEEELVPELQWLHDNPLATLVFKDCINGFIGSIDIDTQGNVLL 149
Query: 581 DWVKKNQIKQILVAGICTDVCVLDFVCSVLSARNRQFLPPLENVIVSTEACSTYDVPLHV 402
DW+ KN+++ ++V GICTD+CV+DFV ++LS RN P L++V V + C+T+D+ +
Sbjct: 150 DWINKNKLEALVVVGICTDICVMDFVVTILSVRNHGLAPTLKDVAVYDQGCATFDMTAQM 209
Query: 401 A--KTNKDWVSHPQELMHHIGLYIACGRGAEIASEV 300
A K HPQ++ HH+GLY RGA IAS +
Sbjct: 210 AAEKGLPKTAIHPQKISHHVGLYTMAERGAFIASTI 245
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 929,801,336
Number of Sequences: 1393205
Number of extensions: 21819778
Number of successful extensions: 54062
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 51228
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 53998
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 58773479151
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)