Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
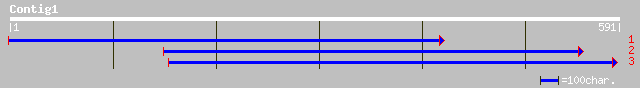
Query= KMC003016A_C01 KMC003016A_c01
(588 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC04594.1| unnamed protein product [Homo sapiens] 38 0.11
ref|ZP_00072522.1| hypothetical protein [Trichodesmium erythraeu... 36 0.41
ref|NP_735796.1| Unknown [Streptococcus agalactiae NEM316] gi|24... 35 0.70
dbj|BAC33062.1| unnamed protein product [Mus musculus] 35 0.92
dbj|BAA31470.1| CXC chemokine receptor-2 [Cyprinus carpio] 33 2.0
>dbj|BAC04594.1| unnamed protein product [Homo sapiens]
Length = 134
Score = 37.7 bits (86), Expect = 0.11
Identities = 17/56 (30%), Positives = 25/56 (44%), Gaps = 3/56 (5%)
Frame = +3
Query: 345 LCCLCLCAICLCI---FCMCISTRYCYFVYTTTNFRAI*NCESATVYGKRLCVFCC 503
+CC+C+C CLC+ C+C+ C +V I C +CVF C
Sbjct: 5 VCCVCMCRWCLCVCIYMCVCVPVCMCVYVCVYVRICVICVC---------VCVFVC 51
Score = 32.0 bits (71), Expect = 5.9
Identities = 19/60 (31%), Positives = 26/60 (42%), Gaps = 8/60 (13%)
Frame = +3
Query: 351 CLCLCA-----ICLCIF---CMCISTRYCYFVYTTTNFRAI*NCESATVYGKRLCVFCCL 506
C+C+C +C C+F C+C+ C FV C VY LCVF C+
Sbjct: 43 CVCVCVFVCAHMCSCVFVYVCICVCLYVCVFV-----------CVCMYVY---LCVFVCM 88
>ref|ZP_00072522.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 302
Score = 35.8 bits (81), Expect = 0.41
Identities = 28/84 (33%), Positives = 37/84 (43%), Gaps = 3/84 (3%)
Frame = -2
Query: 557 ENISRYLVNVWKEK---TRTQATEHT*TLTIDRCTFTVLNCSKICCCINKITVSCRNTHT 387
ENI VN E+ T T TE T T TI+ +KI +K+ + T T
Sbjct: 53 ENIDSENVNSQTEENQVTETLETETTKTTTIETKDAETTTTAKIIYKPSKVNIET-TTST 111
Query: 386 KNTQTDSTETETTEPSPKTQVFKV 315
KN TD TET T+ P ++
Sbjct: 112 KNPATDRTETTTSAQQPTANTNRI 135
>ref|NP_735796.1| Unknown [Streptococcus agalactiae NEM316]
gi|24412939|emb|CAD47018.1| Unknown [Streptococcus
agalactiae NEM316]
Length = 931
Score = 35.0 bits (79), Expect = 0.70
Identities = 22/56 (39%), Positives = 28/56 (49%), Gaps = 3/56 (5%)
Frame = -1
Query: 528 LERKNQNPSNRTHIDA---YHRPLHFHSSKLL*NLLLYKQNNSIL*KYTYKKYTNR 370
LE K+ NP H +A YH P HFH K L L K+ NS +Y Y K ++
Sbjct: 115 LEEKS-NPKFAYHKEADIKYHSPFHFHKEKTLERLQAEKEVNSAKCEYKYLKQAHK 169
>dbj|BAC33062.1| unnamed protein product [Mus musculus]
Length = 105
Score = 34.7 bits (78), Expect = 0.92
Identities = 8/23 (34%), Positives = 16/23 (68%)
Frame = +3
Query: 345 LCCLCLCAICLCIFCMCISTRYC 413
+C +C+C +C+C+ C+C+ C
Sbjct: 7 VCGVCVCGVCVCVVCVCVCVYVC 29
>dbj|BAA31470.1| CXC chemokine receptor-2 [Cyprinus carpio]
Length = 342
Score = 33.5 bits (75), Expect = 2.0
Identities = 18/64 (28%), Positives = 32/64 (49%)
Frame = +1
Query: 328 WVFGEGSVVSVSVLSVCVFFVCVFLQDTVILFIQQQILEQFRTVKVQRSMVSVYVCSVAW 507
W+FG +S L F+ CVFL + + I++ + + +R +V + VC++ W
Sbjct: 91 WMFGTFMCKLISGLQEATFYCCVFLLACISVDRYLAIVKATQFLAQKRHLVGI-VCALVW 149
Query: 508 VLVF 519
V F
Sbjct: 150 VCAF 153
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 470,107,926
Number of Sequences: 1393205
Number of extensions: 9822804
Number of successful extensions: 30731
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 28118
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30310
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)