Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003012A_C01 KMC003012A_c01
(582 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
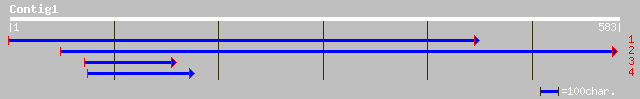
ref|NP_565139.1| expressed protein; protein id: At1g76730.1, sup... 141 5e-33
dbj|BAC43679.1| unknown protein [Arabidopsis thaliana] 140 2e-32
dbj|BAB14739.1| unnamed protein product [Homo sapiens] 58 8e-08
gb|AAH13911.1|AAH13911 Similar to hypothetical protein FLJ12998 ... 58 8e-08
ref|NP_766349.1| hypothetical protein C920004D05 [Mus musculus] ... 58 1e-07
>ref|NP_565139.1| expressed protein; protein id: At1g76730.1, supported by cDNA:
33304. [Arabidopsis thaliana] gi|25406463|pir||G96795
hypothetical protein F28O16.10 [imported] - Arabidopsis
thaliana gi|6143890|gb|AAF04436.1|AC010718_5
hypothetical protein; 41475-39805 [Arabidopsis thaliana]
Length = 354
Score = 141 bits (356), Expect = 5e-33
Identities = 81/136 (59%), Positives = 93/136 (67%)
Frame = -2
Query: 581 VAVDPRTGARLGKGEGFAELEYGMLRYMGAIDDSTQVVTLVHDCPLVDDISS*EGC*SHD 402
VAV+P+TGARLGKGEGFAELEYGMLRYMGAIDDST VVT VHDC LVDDI E HD
Sbjct: 223 VAVNPQTGARLGKGEGFAELEYGMLRYMGAIDDSTPVVTTVHDCQLVDDIPL-EKLAIHD 281
Query: 401 VSS*YQFVLPNPGHFSSRTSIPQACKEFIGTNCLQRTLGQVRILRELKKRIERETGQKLP 222
V + + T IP+ + L Q+RILRELK R+E++TG+KLP
Sbjct: 282 VP--VDIICTPTRVIFTNTPIPKP-QGIYWDKLSPEKLRQIRILRELKNRLEKKTGRKLP 338
Query: 221 TGPSAKLPPTAQRSRR 174
TGPS KLPPTA+R RR
Sbjct: 339 TGPSEKLPPTAERKRR 354
>dbj|BAC43679.1| unknown protein [Arabidopsis thaliana]
Length = 354
Score = 140 bits (352), Expect = 2e-32
Identities = 80/136 (58%), Positives = 92/136 (66%)
Frame = -2
Query: 581 VAVDPRTGARLGKGEGFAELEYGMLRYMGAIDDSTQVVTLVHDCPLVDDISS*EGC*SHD 402
VAV+P+TG RLGKGEGFAELEYGMLRYMGAIDDST VVT VHDC LVDDI E HD
Sbjct: 223 VAVNPQTGVRLGKGEGFAELEYGMLRYMGAIDDSTPVVTTVHDCQLVDDIPL-EKLAIHD 281
Query: 401 VSS*YQFVLPNPGHFSSRTSIPQACKEFIGTNCLQRTLGQVRILRELKKRIERETGQKLP 222
V + + T IP+ + L Q+RILRELK R+E++TG+KLP
Sbjct: 282 VP--VDIICTPTRVIFTNTPIPKP-QGIYWDKLSPEKLRQIRILRELKNRLEKKTGRKLP 338
Query: 221 TGPSAKLPPTAQRSRR 174
TGPS KLPPTA+R RR
Sbjct: 339 TGPSEKLPPTAERKRR 354
>dbj|BAB14739.1| unnamed protein product [Homo sapiens]
Length = 382
Score = 58.2 bits (139), Expect = 8e-08
Identities = 28/48 (58%), Positives = 36/48 (74%)
Frame = -2
Query: 581 VAVDPRTGARLGKGEGFAELEYGMLRYMGAIDDSTQVVTLVHDCPLVD 438
VAV + G R+GKGEG+A+LEY M+ MGA+ T VVT+VHDC +VD
Sbjct: 141 VAVSEK-GWRIGKGEGYADLEYAMMVSMGAVSKETPVVTIVHDCQVVD 187
>gb|AAH13911.1|AAH13911 Similar to hypothetical protein FLJ12998 [Homo sapiens]
Length = 228
Score = 58.2 bits (139), Expect = 8e-08
Identities = 28/48 (58%), Positives = 36/48 (74%)
Frame = -2
Query: 581 VAVDPRTGARLGKGEGFAELEYGMLRYMGAIDDSTQVVTLVHDCPLVD 438
VAV + G R+GKGEG+A+LEY M+ MGA+ T VVT+VHDC +VD
Sbjct: 141 VAVSEK-GWRIGKGEGYADLEYAMMVSMGAVSKETPVVTIVHDCQVVD 187
>ref|NP_766349.1| hypothetical protein C920004D05 [Mus musculus]
gi|26350447|dbj|BAC38863.1| unnamed protein product [Mus
musculus]
Length = 353
Score = 57.8 bits (138), Expect = 1e-07
Identities = 28/48 (58%), Positives = 36/48 (74%)
Frame = -2
Query: 581 VAVDPRTGARLGKGEGFAELEYGMLRYMGAIDDSTQVVTLVHDCPLVD 438
VAV + G R+GKGEG+A+LEY M+ MGA+ T VVT+VHDC +VD
Sbjct: 122 VAVSEK-GWRIGKGEGYADLEYAMMVSMGAVHKGTPVVTIVHDCQVVD 168
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 536,806,005
Number of Sequences: 1393205
Number of extensions: 12216139
Number of successful extensions: 32957
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 31553
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32923
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)