Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003009A_C02 KMC003009A_c02
(551 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
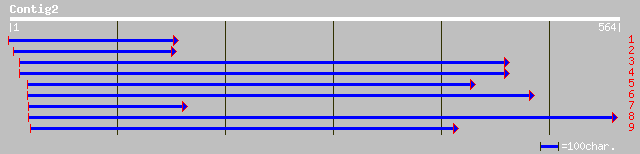
gb|AAM65866.1| unknown [Arabidopsis thaliana] 116 2e-25
ref|NP_197853.1| putative protein; protein id: At5g24650.1, supp... 116 2e-25
ref|NP_190525.1| putative protein; protein id: At3g49560.1, supp... 110 9e-24
ref|NP_177175.2| hypothetical protein; protein id: At1g70180.1, ... 36 0.35
gb|AAH46229.1| Similar to Sec23-interacting protein p125 [Mus mu... 34 1.0
>gb|AAM65866.1| unknown [Arabidopsis thaliana]
Length = 246
Score = 116 bits (290), Expect = 2e-25
Identities = 58/88 (65%), Positives = 66/88 (74%)
Frame = -1
Query: 551 TSGLFFALVQGGLFPLGPKFSQPPVEDTNYVKTRHMLHNLGLQNYEKHFKKGLLTDNTLP 372
T+ FA+ QG F LG +FS+P VED Y + R ML LGL+ YEK+FKKGLL D TLP
Sbjct: 149 TTAAGFAVFQGVFFKLGERFSKPSVEDPYYTRGRSMLLKLGLEKYEKNFKKGLLADPTLP 208
Query: 371 LLTDRALRDVKIPPGPRLLILDHIPREP 288
LLTD ALRDV IPPGPRLLILDHI R+P
Sbjct: 209 LLTDSALRDVSIPPGPRLLILDHIQRDP 236
>ref|NP_197853.1| putative protein; protein id: At5g24650.1, supported by cDNA:
5684., supported by cDNA: gi_17979478 [Arabidopsis
thaliana] gi|10177865|dbj|BAB11217.1|
emb|CAB62460.1~gene_id:K18P6.19~similar to unknown
protein [Arabidopsis thaliana]
gi|17979479|gb|AAL50076.1| AT5g24650/K18P6_19
[Arabidopsis thaliana] gi|22655440|gb|AAM98312.1|
At5g24650/K18P6_19 [Arabidopsis thaliana]
Length = 259
Score = 116 bits (290), Expect = 2e-25
Identities = 58/88 (65%), Positives = 66/88 (74%)
Frame = -1
Query: 551 TSGLFFALVQGGLFPLGPKFSQPPVEDTNYVKTRHMLHNLGLQNYEKHFKKGLLTDNTLP 372
T+ FA+ QG F LG +FS+P VED Y + R ML LGL+ YEK+FKKGLL D TLP
Sbjct: 162 TTAAGFAVFQGVFFKLGERFSKPSVEDPYYTRGRSMLLKLGLEKYEKNFKKGLLADPTLP 221
Query: 371 LLTDRALRDVKIPPGPRLLILDHIPREP 288
LLTD ALRDV IPPGPRLLILDHI R+P
Sbjct: 222 LLTDSALRDVSIPPGPRLLILDHIQRDP 249
>ref|NP_190525.1| putative protein; protein id: At3g49560.1, supported by cDNA:
15698., supported by cDNA: gi_13430557 [Arabidopsis
thaliana] gi|11285728|pir||T46233 hypothetical protein
T9C5.150 - Arabidopsis thaliana
gi|6561956|emb|CAB62460.1| putative protein [Arabidopsis
thaliana] gi|13430558|gb|AAK25901.1|AF360191_1 unknown
protein [Arabidopsis thaliana]
gi|21553682|gb|AAM62775.1| unknown [Arabidopsis
thaliana] gi|25054947|gb|AAN71950.1| unknown protein
[Arabidopsis thaliana]
Length = 261
Score = 110 bits (276), Expect = 9e-24
Identities = 54/88 (61%), Positives = 65/88 (73%)
Frame = -1
Query: 551 TSGLFFALVQGGLFPLGPKFSQPPVEDTNYVKTRHMLHNLGLQNYEKHFKKGLLTDNTLP 372
T+ FA+ QG F LG +FS+P ED + + R ML LGL+ YEK+FKKGLLTD TLP
Sbjct: 167 TTAAGFAVFQGVFFKLGERFSKPSTEDPFFTRGRTMLVKLGLEKYEKNFKKGLLTDPTLP 226
Query: 371 LLTDRALRDVKIPPGPRLLILDHIPREP 288
LLTD AL+D IPPGPRL+ILDHI R+P
Sbjct: 227 LLTDSALKDANIPPGPRLMILDHIQRDP 254
>ref|NP_177175.2| hypothetical protein; protein id: At1g70180.1, supported by cDNA:
gi_17473773 [Arabidopsis thaliana]
gi|17473774|gb|AAL38323.1| unknown protein [Arabidopsis
thaliana]
Length = 460
Score = 35.8 bits (81), Expect = 0.35
Identities = 22/74 (29%), Positives = 33/74 (43%)
Frame = -1
Query: 509 PLGPKFSQPPVEDTNYVKTRHMLHNLGLQNYEKHFKKGLLTDNTLPLLTDRALRDVKIPP 330
PLG P + L+++GL Y FK+ + T+ + + L+D+ IP
Sbjct: 385 PLGSIMQNSPFSVDEPLSVDGFLNSIGLGKYSLAFKREEVDMTTIKQMKESDLKDLIIPM 444
Query: 329 GPRLLILDHIPREP 288
GPR IL I P
Sbjct: 445 GPRKKILQAIASLP 458
>gb|AAH46229.1| Similar to Sec23-interacting protein p125 [Mus musculus]
Length = 706
Score = 34.3 bits (77), Expect = 1.0
Identities = 19/47 (40%), Positives = 26/47 (54%)
Frame = -1
Query: 443 LHNLGLQNYEKHFKKGLLTDNTLPLLTDRALRDVKIPPGPRLLILDH 303
L L L + F+K + L L TDR L+++ IP GPR IL+H
Sbjct: 401 LKKLQLSEFVTVFEKEKVDREALALCTDRDLQEMGIPLGPRKKILNH 447
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 455,774,273
Number of Sequences: 1393205
Number of extensions: 9303757
Number of successful extensions: 24216
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 23321
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24182
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19234190289
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)