Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
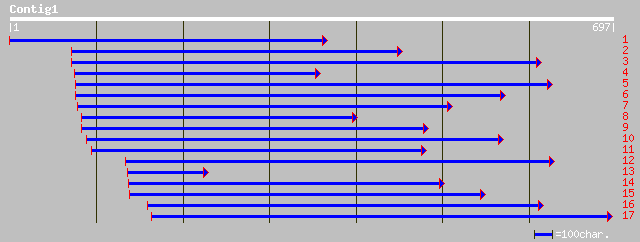
Query= KMC002997A_C01 KMC002997A_c01
(688 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565590.1| expressed protein; protein id: At2g25280.1, sup... 144 1e-33
gb|EAA13930.1| agCP8751 [Anopheles gambiae str. PEST] 108 4e-24
ref|NP_650252.1| CG8031-PA [Drosophila melanogaster] gi|7299707|... 109 5e-24
ref|XP_216638.1| similar to RIKEN cDNA 0610016J10 [Mus musculus]... 111 1e-23
ref|NP_598532.1| RIKEN cDNA 0610016J10 [Mus musculus] gi|1548946... 111 1e-23
>ref|NP_565590.1| expressed protein; protein id: At2g25280.1, supported by cDNA:
25127., supported by cDNA: gi_16604562, supported by
cDNA: gi_20259160 [Arabidopsis thaliana]
gi|25412293|pir||E84646 hypothetical protein At2g25280
[imported] - Arabidopsis thaliana
gi|4567249|gb|AAD23663.1| expressed protein [Arabidopsis
thaliana] gi|16604563|gb|AAL24083.1| unknown protein
[Arabidopsis thaliana] gi|20259161|gb|AAM14296.1|
unknown protein [Arabidopsis thaliana]
gi|21554336|gb|AAM63443.1| unknown [Arabidopsis
thaliana]
Length = 291
Score = 144 bits (362), Expect = 1e-33
Identities = 72/123 (58%), Positives = 89/123 (71%), Gaps = 1/123 (0%)
Frame = -3
Query: 686 PTNFFSISSDFCHWGARFNYQHYDKKHGPIYKSIEALDKMGMDIIEIGDPDSFKRYLSEY 507
P NFFS+SSDFCHWG+RFNY HYD HG I+KSIEALDK GMDIIE GDPD+FK+YL E+
Sbjct: 176 PKNFFSVSSDFCHWGSRFNYMHYDNTHGAIHKSIEALDKKGMDIIETGDPDAFKKYLLEF 235
Query: 506 DNTICGRHPISVFLHLLAILPYAEELLNQDQNKISSL*AVQPMP-KLQGIAVLSYASAAA 330
+NTICGRHPIS+FLH+L + + KI+ L Q + + +SYASAAA
Sbjct: 236 ENTICGRHPISIFLHMLK--------HSSSKIKINFLRYEQSSQCQTMRDSSVSYASAAA 287
Query: 329 KID 321
K++
Sbjct: 288 KLE 290
>gb|EAA13930.1| agCP8751 [Anopheles gambiae str. PEST]
Length = 330
Score = 108 bits (270), Expect(2) = 4e-24
Identities = 48/74 (64%), Positives = 55/74 (73%)
Frame = -3
Query: 686 PTNFFSISSDFCHWGARFNYQHYDKKHGPIYKSIEALDKMGMDIIEIGDPDSFKRYLSEY 507
P N F ISSDFCHWG RF Y +Y++ GPIYK IE LDKMGMD+IE DSF YL +Y
Sbjct: 213 PQNLFVISSDFCHWGQRFRYTYYEEGSGPIYKWIETLDKMGMDLIETLKADSFSEYLRKY 272
Query: 506 DNTICGRHPISVFL 465
+NTICGRHPI V +
Sbjct: 273 NNTICGRHPIGVLM 286
Score = 25.0 bits (53), Expect(2) = 4e-24
Identities = 9/29 (31%), Positives = 17/29 (58%)
Frame = -1
Query: 463 IFLQFYHMLRNCSTKIKIKFLRYEQSSPC 377
+ +Q L+ ++ KFL+Y+QS+ C
Sbjct: 284 VLMQAVEELKRRGHRMSFKFLKYDQSNQC 312
>ref|NP_650252.1| CG8031-PA [Drosophila melanogaster] gi|7299707|gb|AAF54889.1|
CG8031-PA [Drosophila melanogaster]
gi|16769746|gb|AAL29092.1| LP04475p [Drosophila
melanogaster]
Length = 295
Score = 109 bits (272), Expect(2) = 5e-24
Identities = 50/74 (67%), Positives = 56/74 (75%)
Frame = -3
Query: 686 PTNFFSISSDFCHWGARFNYQHYDKKHGPIYKSIEALDKMGMDIIEIGDPDSFKRYLSEY 507
PTN F ISSDFCHWG RF+Y +YD G I+KSIE LDK GMDIIE +P SF YL +Y
Sbjct: 176 PTNLFVISSDFCHWGHRFSYTYYDSSCGAIHKSIEKLDKQGMDIIESLNPHSFTEYLRKY 235
Query: 506 DNTICGRHPISVFL 465
+NTICGRHPI V L
Sbjct: 236 NNTICGRHPIGVML 249
Score = 23.9 bits (50), Expect(2) = 5e-24
Identities = 9/15 (60%), Positives = 11/15 (73%)
Frame = -1
Query: 421 KIKIKFLRYEQSSPC 377
K+ KFL+Y QSS C
Sbjct: 262 KMSFKFLKYAQSSQC 276
>ref|XP_216638.1| similar to RIKEN cDNA 0610016J10 [Mus musculus] [Rattus norvegicus]
Length = 180
Score = 111 bits (277), Expect = 1e-23
Identities = 50/80 (62%), Positives = 58/80 (72%)
Frame = -3
Query: 686 PTNFFSISSDFCHWGARFNYQHYDKKHGPIYKSIEALDKMGMDIIEIGDPDSFKRYLSEY 507
P+N F +SSDFCHWG RF Y +YD+ G IY+SIE LDKMGM IIE DP SF YL +Y
Sbjct: 63 PSNLFVVSSDFCHWGQRFRYSYYDESQGEIYRSIEHLDKMGMSIIEQLDPVSFSNYLKKY 122
Query: 506 DNTICGRHPISVFLHLLAIL 447
NTICGRHPI V L+ + L
Sbjct: 123 HNTICGRHPIGVLLNAITEL 142
>ref|NP_598532.1| RIKEN cDNA 0610016J10 [Mus musculus] gi|15489467|gb|AAH13819.1|
RIKEN cDNA 0610016J10 gene [Mus musculus]
gi|17046305|gb|AAL34463.1|AF363447_1 C21orf19-like
protein [Mus musculus]
Length = 297
Score = 111 bits (277), Expect = 1e-23
Identities = 50/80 (62%), Positives = 58/80 (72%)
Frame = -3
Query: 686 PTNFFSISSDFCHWGARFNYQHYDKKHGPIYKSIEALDKMGMDIIEIGDPDSFKRYLSEY 507
P+N F +SSDFCHWG RF Y +YD+ G IY+SIE LDKMGM IIE DP SF YL +Y
Sbjct: 180 PSNLFVVSSDFCHWGQRFRYSYYDESQGEIYRSIEHLDKMGMSIIEQLDPVSFSNYLKKY 239
Query: 506 DNTICGRHPISVFLHLLAIL 447
NTICGRHPI V L+ + L
Sbjct: 240 HNTICGRHPIGVLLNAITEL 259
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 528,244,518
Number of Sequences: 1393205
Number of extensions: 10726277
Number of successful extensions: 22065
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 21556
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22052
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30835865868
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)