Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002994A_C01 KMC002994A_c01
(502 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
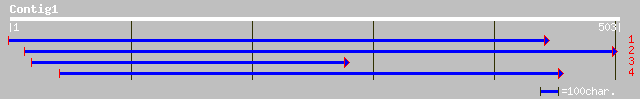
dbj|BAA97502.1| myosin heavy chain-like [Arabidopsis thaliana] 127 5e-29
ref|NP_200829.1| putative protein; protein id: At5g60210.1 [Arab... 127 5e-29
emb|CAC84774.1| P70 protein [Nicotiana tabacum] 120 9e-27
ref|NP_181245.1| putative myosin heavy chain; protein id: At2g37... 106 2e-22
dbj|BAB92927.1| P0460C04.20 [Oryza sativa (japonica cultivar-gro... 82 3e-15
>dbj|BAA97502.1| myosin heavy chain-like [Arabidopsis thaliana]
Length = 564
Score = 127 bits (320), Expect = 5e-29
Identities = 65/95 (68%), Positives = 76/95 (79%)
Frame = -2
Query: 498 RVTEQLEAAQASNSEMEAELRRMKVQSDQWRKAAEAAAAMLSAGNNGKITERTMSLDDNS 319
RVTEQLEA QASNSEME ELR++KVQS+QWRKAAEAA AMLSAGNNGK E +
Sbjct: 476 RVTEQLEATQASNSEMETELRKLKVQSNQWRKAAEAATAMLSAGNNGKFAENYNQTN--- 532
Query: 318 YKCSPFAEENDNDYLRKKNGNMLKKIGVLWKKPQK 214
SP++E+ D++ +KKNGN+LKKIGVLWKKPQK
Sbjct: 533 ---SPYSEDIDDELTKKKNGNVLKKIGVLWKKPQK 564
>ref|NP_200829.1| putative protein; protein id: At5g60210.1 [Arabidopsis thaliana]
Length = 629
Score = 127 bits (320), Expect = 5e-29
Identities = 65/95 (68%), Positives = 76/95 (79%)
Frame = -2
Query: 498 RVTEQLEAAQASNSEMEAELRRMKVQSDQWRKAAEAAAAMLSAGNNGKITERTMSLDDNS 319
RVTEQLEA QASNSEME ELR++KVQS+QWRKAAEAA AMLSAGNNGK E +
Sbjct: 500 RVTEQLEATQASNSEMETELRKLKVQSNQWRKAAEAATAMLSAGNNGKFAENYNQTN--- 556
Query: 318 YKCSPFAEENDNDYLRKKNGNMLKKIGVLWKKPQK 214
SP++E+ D++ +KKNGN+LKKIGVLWKKPQK
Sbjct: 557 ---SPYSEDIDDELTKKKNGNVLKKIGVLWKKPQK 588
>emb|CAC84774.1| P70 protein [Nicotiana tabacum]
Length = 601
Score = 120 bits (301), Expect = 9e-27
Identities = 65/99 (65%), Positives = 78/99 (78%), Gaps = 3/99 (3%)
Frame = -2
Query: 501 ARVTEQLEAAQASNSEMEAELRRMKVQSDQWRKAAEAAAAMLSAGNNGKITERTMSLDDN 322
ARVTE+L+AAQ +NSEME ELRR+KVQ DQWRKAAEAAA+MLSAGNNGK ERT SLD +
Sbjct: 503 ARVTEELDAAQTANSEMETELRRLKVQCDQWRKAAEAAASMLSAGNNGKYVERTGSLDYH 562
Query: 321 SY---KCSPFAEENDNDYLRKKNGNMLKKIGVLWKKPQK 214
+ SP ++ ++D +KKN NMLK+IGVL KK QK
Sbjct: 563 TIGGKLGSPLLDDLEDDSPKKKNNNMLKRIGVLLKKGQK 601
>ref|NP_181245.1| putative myosin heavy chain; protein id: At2g37080.1 [Arabidopsis
thaliana] gi|25408529|pir||C84788 probable myosin heavy
chain [imported] - Arabidopsis thaliana
gi|4371293|gb|AAD18151.1| putative myosin heavy chain
[Arabidopsis thaliana]
Length = 583
Score = 106 bits (264), Expect = 2e-22
Identities = 62/104 (59%), Positives = 73/104 (69%), Gaps = 11/104 (10%)
Frame = -2
Query: 492 TEQLEAAQASNSEMEAELRRMKVQSDQWRKAAEAAAAMLSAGN-----NGKITERTMSLD 328
TEQL AAQ +N+E+EAELRR+KVQ DQWRKAAEAAA MLS GN NGK ERT SL+
Sbjct: 480 TEQLGAAQVTNTELEAELRRLKVQCDQWRKAAEAAATMLSGGNNNNNSNGKYVERTGSLE 539
Query: 327 D----NSYKCSPFAEENDNDYL--RKKNGNMLKKIGVLWKKPQK 214
+ SP+ E D++ +KKNG+MLKKIGVL KK QK
Sbjct: 540 SPLRRRNVNMSPYMGETDDELSSPKKKNGSMLKKIGVLLKKSQK 583
>dbj|BAB92927.1| P0460C04.20 [Oryza sativa (japonica cultivar-group)]
Length = 947
Score = 82.0 bits (201), Expect = 3e-15
Identities = 45/100 (45%), Positives = 66/100 (66%), Gaps = 7/100 (7%)
Frame = -2
Query: 501 ARVTEQLEAAQ--ASNSEMEAELRRMKVQSDQWRKAAEAAAAMLSA--GNNGKITERTMS 334
A+ T+ +AA A EMEAELRR++VQ++QWRKAAE A A+L+ G NGK+ +R+ S
Sbjct: 848 AKATDAADAAAEAAKKGEMEAELRRLRVQAEQWRKAAETAMALLTVGKGGNGKVVDRSES 907
Query: 333 LDDNSYKCSPFA---EENDNDYLRKKNGNMLKKIGVLWKK 223
L+ +A +E D+D +KNGN+L++I +WKK
Sbjct: 908 LEGGGGGGGKYAGLWDELDDDAAARKNGNVLRRISGMWKK 947
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 423,659,281
Number of Sequences: 1393205
Number of extensions: 8955397
Number of successful extensions: 25807
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 24783
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25763
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 15072921604
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)