Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002978A_C01 KMC002978A_c01
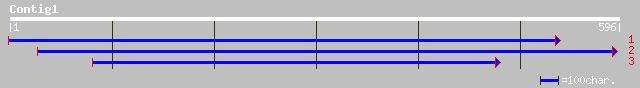
(595 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO50779.1| similar to Mus musculus (Mouse). Sex-determining ... 45 0.001
gb|AAL17914.1| homeobox protein hox4x [Petromyzon marinus] 42 0.008
sp|P36417|GBF_DICDI G-BOX BINDING FACTOR (GBF) gi|1076833|pir||A... 41 0.010
ref|NP_066232.2| NADH dehydrogenase subunit 4 [Taenia crassiceps... 41 0.013
gb|ZP_00026734.1| hypothetical protein [Ralstonia metallidurans] 41 0.013
>gb|AAO50779.1| similar to Mus musculus (Mouse). Sex-determining region Y protein
(Testis-determining factor) [Dictyostelium discoideum]
Length = 1461
Score = 44.7 bits (104), Expect = 0.001
Identities = 31/155 (20%), Positives = 58/155 (37%), Gaps = 22/155 (14%)
Frame = +1
Query: 193 HQNKESTNTMKSQQTS*CPHVTP*PKRKEHQHDHSNRQDE-----HW*PQIDHRHRTGRN 357
HQ + QQ PH P +++ HQ++H Q H Q+ + + +
Sbjct: 947 HQQPHQHQQQQHQQ----PHQQPHHQQQHHQNNHQQHQQHQQHQHHQQQQLQQQQQQQQQ 1002
Query: 358 QDCH*HEVEKHPNDPRPLEKERNQVKGYEKQQQE-----------------HEESK*HQP 486
Q H H+ H + ++++ Q + ++QQQ+ H++ HQ
Sbjct: 1003 QQHHQHQQHHHHQQQQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQQQQHQHQQQHQHQQ 1062
Query: 487 HRHQ*IYSYGNPHLTQFSNDPNNSYQCDRHHDQSN 591
H+HQ H Q + + +Q H +
Sbjct: 1063 HQHQQHQHQHQQHQHQHQHQHQHQHQHQHQHQHQH 1097
Score = 42.0 bits (97), Expect = 0.006
Identities = 24/136 (17%), Positives = 57/136 (41%), Gaps = 4/136 (2%)
Frame = +1
Query: 190 KHQNKESTNTMKSQQTS*CPHVTP*PKRKEHQHD----HSNRQDEHW*PQIDHRHRTGRN 357
+HQ+ + + QQ +++ HQH H +Q + Q + + +
Sbjct: 984 QHQHHQQQQLQQQQQQQ--------QQQQHHQHQQHHHHQQQQQQQQQQQQQQQQQQQQQ 1035
Query: 358 QDCH*HEVEKHPNDPRPLEKERNQVKGYEKQQQEHEESK*HQPHRHQ*IYSYGNPHLTQF 537
Q H+ ++ + ++++Q + ++ QQ +H+ + H+HQ + + + H Q
Sbjct: 1036 QQQQQHQQQQQQQQQQHQHQQQHQHQQHQHQQHQHQHQQHQHQHQHQHQHQHQHQHQHQH 1095
Query: 538 SNDPNNSYQCDRHHDQ 585
+ + +Q H Q
Sbjct: 1096 QHQHQHQHQHQHQHQQ 1111
Score = 42.0 bits (97), Expect = 0.006
Identities = 18/78 (23%), Positives = 39/78 (49%), Gaps = 1/78 (1%)
Frame = +1
Query: 268 KRKEHQHDHSNRQDEHW*PQIDHRHRTGRNQDCH*HEVEKHPNDPRPLEKERNQVKGYEK 447
++++HQH ++ +H Q H+H+ ++Q H H+ + + + ++
Sbjct: 1048 QQQQHQHQQQHQHQQHQHQQHQHQHQQHQHQHQHQHQHQHQHQHQHQHQHQHQHQHQHQH 1107
Query: 448 QQQEHEESK*H-QPHRHQ 498
Q Q+H++ + H Q H HQ
Sbjct: 1108 QHQQHQQHQQHQQQHHHQ 1125
Score = 41.6 bits (96), Expect = 0.008
Identities = 27/134 (20%), Positives = 57/134 (42%), Gaps = 1/134 (0%)
Frame = +1
Query: 193 HQNKESTNTMKSQQTS*CPHVTP*PKRKEHQHDHSNRQDEHW*PQIDHRHRTGRNQDC-H 369
H +++ + QQ ++++HQ +Q +H Q H+H+ ++Q H
Sbjct: 1012 HHHQQQQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQQQQHQHQQ-QHQHQQHQHQQHQH 1070
Query: 370 *HEVEKHPNDPRPLEKERNQVKGYEKQQQEHEESK*HQPHRHQ*IYSYGNPHLTQFSNDP 549
H+ +H + + + ++Q + + Q +H+ HQ +HQ + H
Sbjct: 1071 QHQQHQHQHQHQHQHQHQHQHQHQHQHQHQHQHQHQHQHQQHQQHQQHQQQH-------- 1122
Query: 550 NNSYQCDRHHDQSN 591
+Q ++HH Q N
Sbjct: 1123 --HHQQNQHHHQQN 1134
Score = 40.8 bits (94), Expect = 0.013
Identities = 27/132 (20%), Positives = 51/132 (38%)
Frame = +1
Query: 190 KHQNKESTNTMKSQQTS*CPHVTP*PKRKEHQHDHSNRQDEHW*PQIDHRHRTGRNQDCH 369
+ Q ++ + QQ H +++ HQ H ++Q +H P H+ +Q+ H
Sbjct: 917 QQQQQQQQQQPQPQQLQLPQHQHQQQQQQPHQQPHQHQQQQHQQPHQQPHHQQQHHQNNH 976
Query: 370 *HEVEKHPNDPRPLEKERNQVKGYEKQQQEHEESK*HQPHRHQ*IYSYGNPHLTQFSNDP 549
+ ++H +++ Q + ++QQQ H+ HQ H H Q
Sbjct: 977 -QQHQQHQQHQHHQQQQLQQQQQQQQQQQHHQ----HQQHHHHQQQQQQQQQQQQQQQQQ 1031
Query: 550 NNSYQCDRHHDQ 585
Q + H Q
Sbjct: 1032 QQQQQQQQQHQQ 1043
Score = 40.0 bits (92), Expect = 0.022
Identities = 26/144 (18%), Positives = 55/144 (38%), Gaps = 9/144 (6%)
Frame = +1
Query: 181 PKVKHQNKESTNTMKSQQTS*CPHVTP*PKRKEHQHDHSNRQDE---------HW*PQID 333
P+ +HQ ++ + Q H P + Q H N + H Q+
Sbjct: 935 PQHQHQQQQQQPHQQPHQHQQQQHQQPHQQPHHQQQHHQNNHQQHQQHQQHQHHQQQQLQ 994
Query: 334 HRHRTGRNQDCH*HEVEKHPNDPRPLEKERNQVKGYEKQQQEHEESK*HQPHRHQ*IYSY 513
+ + + Q H H+ H + ++++ Q + ++QQQ+ ++ Q + Q + +
Sbjct: 995 QQQQQQQQQQHHQHQQHHHHQQQQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQQQQHQH 1054
Query: 514 GNPHLTQFSNDPNNSYQCDRHHDQ 585
H Q + +Q +H Q
Sbjct: 1055 QQQHQHQQHQHQQHQHQHQQHQHQ 1078
Score = 38.5 bits (88), Expect = 0.065
Identities = 24/104 (23%), Positives = 49/104 (47%), Gaps = 1/104 (0%)
Frame = +1
Query: 190 KHQNKESTNTMKSQQTS*CPHVTP*PKRKEHQHDHSNRQDEHW*PQIDHRHRTGRNQDCH 369
+HQ ++ + Q H + ++HQH H Q +H Q H+H+ ++Q H
Sbjct: 1040 QHQQQQQQQQQQHQHQQ--QHQHQQHQHQQHQHQHQQHQHQH---QHQHQHQH-QHQHQH 1093
Query: 370 *HEVEKHPNDPRPLEKERNQVKGYEKQQQEHEESK-*HQPHRHQ 498
H+ + + +++Q +QQ H++++ HQ ++HQ
Sbjct: 1094 QHQHQHQHQHQHQHQHQQHQQHQQHQQQHHHQQNQHHHQQNQHQ 1137
>gb|AAL17914.1| homeobox protein hox4x [Petromyzon marinus]
Length = 381
Score = 41.6 bits (96), Expect = 0.008
Identities = 32/136 (23%), Positives = 53/136 (38%), Gaps = 6/136 (4%)
Frame = +1
Query: 190 KHQNKESTNTMKSQQTS*CPHVTP*PKRKEHQHDHSNRQDEHW*PQIDHRHRTGRNQDCH 369
+H N+++ + + Q+ H + H H H + QD H H H+ R D H
Sbjct: 82 RHLNRQNHHHQQQQEQ----HHQQQHHHQNHNHHHGHHQDHHQ----GHHHQ--RQHDAH 131
Query: 370 *-HEVEKHPNDPRPLEKERNQVKGYEKQQQEHEESK*HQ-----PHRHQ*IYSYGNPHLT 531
H+++ P+ + + +Q + + QQ +H HQ H HQ Y
Sbjct: 132 RDHQLDVLPDCDQQQHHDHHQQQHKQHQQHQHNNHHHHQLHQQQQHHHQQHYKLQQQQQQ 191
Query: 532 QFSNDPNNSYQCDRHH 579
Q Y C+R H
Sbjct: 192 QQQASQAPIYSCERAH 207
>sp|P36417|GBF_DICDI G-BOX BINDING FACTOR (GBF) gi|1076833|pir||A53185 G-box-binding
factor - slime mold (Dictyostelium discoideum)
gi|456562|gb|AAA21021.1| G-box binding factor
Length = 708
Score = 41.2 bits (95), Expect = 0.010
Identities = 25/110 (22%), Positives = 55/110 (49%), Gaps = 3/110 (2%)
Frame = +1
Query: 274 KEHQHDHSNRQDEHW*PQIDHRHRTGRNQDCH*HEVEKHPNDPRPLEKERNQVKGYEKQQ 453
++HQH H +Q + Q H+ + + Q H + + H + + + NQ + ++ Q
Sbjct: 187 QQHQHQHQQQQQQQQHQQQHHQQQQQQQQQHH-QQQQHHQHSQPQQQHQHNQQQQHQHNQ 245
Query: 454 QEHEESK*H---QPHRHQ*IYSYGNPHLTQFSNDPNNSYQCDRHHDQSNS 594
Q+H++ + P + Q + + GN + +N+ NNS + +++ +NS
Sbjct: 246 QQHQQQQNQIQMVPQQPQSLSNSGNNN--NNNNNNNNSNNNNNNNNNNNS 293
Score = 37.7 bits (86), Expect = 0.11
Identities = 18/75 (24%), Positives = 42/75 (56%)
Frame = +1
Query: 271 RKEHQHDHSNRQDEHW*PQIDHRHRTGRNQDCH*HEVEKHPNDPRPLEKERNQVKGYEKQ 450
+++ QH +Q +H Q+ H H+ ++Q H + ++ + + ++++ Q + + +Q
Sbjct: 163 QQQQQHHQQMQQQQHH-QQMQH-HQLQQHQHQHQQQQQQQQHQQQHHQQQQQQQQQHHQQ 220
Query: 451 QQEHEESK*HQPHRH 495
QQ H+ S+ Q H+H
Sbjct: 221 QQHHQHSQPQQQHQH 235
Score = 36.6 bits (83), Expect = 0.25
Identities = 24/109 (22%), Positives = 50/109 (45%)
Frame = +1
Query: 268 KRKEHQHDHSNRQDEHW*PQIDHRHRTGRNQDCH*HEVEKHPNDPRPLEKERNQVKGYEK 447
++ +HQH +Q +H Q H + + Q H H+ ++H +P ++ + + +
Sbjct: 187 QQHQHQHQQQQQQQQH---QQQHHQQQQQQQQQH-HQQQQHHQHSQPQQQHQ-----HNQ 237
Query: 448 QQQEHEESK*HQPHRHQ*IYSYGNPHLTQFSNDPNNSYQCDRHHDQSNS 594
QQQ + HQ ++Q P SN NN+ + +++ +N+
Sbjct: 238 QQQHQHNQQQHQQQQNQIQMVPQQPQ--SLSNSGNNNNNNNNNNNSNNN 284
Score = 34.7 bits (78), Expect = 0.94
Identities = 37/186 (19%), Positives = 68/186 (35%), Gaps = 14/186 (7%)
Frame = +1
Query: 79 NEVQTTSGHYYSYPNQTQSTTR*LTMNP*LNYK*PKVKHQNKESTNTMKSQQTS*CPHVT 258
N + G Y PN NP NY+ ++ +++ QQ H
Sbjct: 83 NVFPSHDGQYPDMPNMVDQYQIHPNQNPHYNYQ-YQLMFMQQQAQQNQPPQQNQQQQHHQ 141
Query: 259 P*PKRKEHQHDHSNRQDEHW*PQIDHRHRTGRNQDCH*-----HEVEKHPNDPRPLEKER 423
++ +H H +Q H Q +H Q H H++++H + + ++++
Sbjct: 142 QQQQQPQH-HQQMQQQQHHQQMQQQQQHHQQMQQQQHHQQMQHHQLQQHQHQHQQQQQQQ 200
Query: 424 NQVKGYEKQQQE-----HEESK*HQ----PHRHQ*IYSYGNPHLTQFSNDPNNSYQCDRH 576
+ + +QQQ+ H++ + HQ +HQ + H Q N Q
Sbjct: 201 QHQQQHHQQQQQQQQQHHQQQQHHQHSQPQQQHQHNQQQQHQHNQQQHQQQQNQIQMVPQ 260
Query: 577 HDQSNS 594
QS S
Sbjct: 261 QPQSLS 266
>ref|NP_066232.2| NADH dehydrogenase subunit 4 [Taenia crassiceps]
gi|12248322|gb|AAG13172.2| NADH dehydrogenase subunit 4
[Taenia crassiceps]
Length = 417
Score = 40.8 bits (94), Expect = 0.013
Identities = 29/107 (27%), Positives = 51/107 (47%), Gaps = 4/107 (3%)
Frame = -2
Query: 480 MLLRFLVLLLLLLVPLHLIPLLFKWSWI----VWMLFNFMLMTVLIPSGPMSVINLWLPM 313
+L+ L L+L+LL ++ + SW V+++ ++L V P+ + WLP+
Sbjct: 132 LLITSLPLILILLYLSYVNNSFYFSSWYAGDDVYLIVYYLLSFVFFTKVPLVPFHTWLPI 191
Query: 312 LILSI*MIVLMFFPLRLGGYMRALTSLLAFHCVCAFFVLMFDLWLFV 172
+ IV +F L GY+ L L + C C F + F +LF+
Sbjct: 192 VHAEATSIVSIF----LSGYIMKLGLLGVYRCSCFIFNVSFLWYLFI 234
>gb|ZP_00026734.1| hypothetical protein [Ralstonia metallidurans]
Length = 101
Score = 40.8 bits (94), Expect = 0.013
Identities = 32/91 (35%), Positives = 53/91 (58%), Gaps = 3/91 (3%)
Frame = -2
Query: 525 MRIPVAINSLMTMRLMLLRFLVLLLLLLVPLHLIPLLFKWSWIVWMLFNFMLMTVLIPSG 346
+R+P+ ++ L +RL LL L+ LL L + +HL+ L + +V +L F+ +T+LI
Sbjct: 7 LRLPLLVHLLPLLRLPLLVHLLPLLRLPLLVHLLLTLLRLPLLVHLL-AFLRLTLLIELL 65
Query: 345 PMSVINLWLPMLILS---I*MIVLMFFPLRL 262
P+ V+ L L L+L+ I + L FF L L
Sbjct: 66 PLLVLRLSLQRLLLTLLEIRRLPLAFFRLAL 96
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 461,031,725
Number of Sequences: 1393205
Number of extensions: 9728714
Number of successful extensions: 39239
Number of sequences better than 10.0: 162
Number of HSP's better than 10.0 without gapping: 33328
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37325
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22854740960
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)