Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
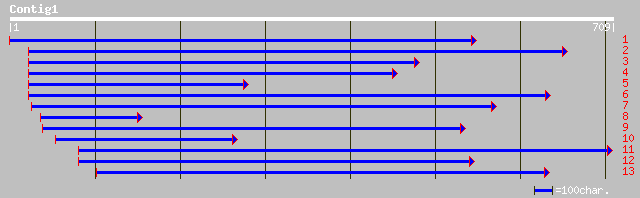
Query= KMC002972A_C01 KMC002972A_c01
(601 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|Q07176|MMK1_MEDSA MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG MM... 207 1e-52
sp|Q06060|MAPK_PEA MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG D5 g... 198 4e-50
gb|AAF65766.1|AF242308_1 mitogen-activated protein kinase [Eupho... 191 5e-48
gb|AAB58396.1| salicylic acid-activated MAP kinase [Nicotiana ta... 190 1e-47
sp|Q40532|NTF4_TOBAC MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG NT... 189 2e-47
>sp|Q07176|MMK1_MEDSA MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG MMK1 (MAP KINASE MSK7)
(MAP KINASE ERK1) gi|1076503|pir||S48123
mitogen-activated protein kinase 7 (EC 2.7.1.-) -
alfalfa gi|289125|gb|AAB41548.1| MAP kinase [Medicago
sativa] gi|298019|emb|CAA47099.1| MAP Kinase [Medicago
sativa]
Length = 387
Score = 207 bits (526), Expect = 1e-52
Identities = 99/103 (96%), Positives = 101/103 (97%)
Frame = -1
Query: 589 GFLNENAKRYIRQLPPYRRQSFQEKFPQVHPEAIDLVEKMLTFDPRKRITVEEALAHPYL 410
GFLNENAKRYIRQLPPYRRQSFQEKFP VHPEAIDLVEKMLTFDPRKRITVE+ALAHPYL
Sbjct: 281 GFLNENAKRYIRQLPPYRRQSFQEKFPHVHPEAIDLVEKMLTFDPRKRITVEDALAHPYL 340
Query: 409 TSLHDISDEPVCMTPFSFDFEQHALSEEQMKELIYREALAFKP 281
TSLHDISDEPVCMTPFSFDFEQHAL+EEQMKELIYREALAF P
Sbjct: 341 TSLHDISDEPVCMTPFSFDFEQHALTEEQMKELIYREALAFNP 383
>sp|Q06060|MAPK_PEA MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG D5 gi|479389|pir||S33635
mitogen-activated protein kinase homolog (clone D5) -
garden pea gi|20808|emb|CAA50036.1| MAP kinase homologue
[Pisum sativum]
Length = 394
Score = 198 bits (504), Expect = 4e-50
Identities = 96/103 (93%), Positives = 98/103 (94%)
Frame = -1
Query: 589 GFLNENAKRYIRQLPPYRRQSFQEKFPQVHPEAIDLVEKMLTFDPRKRITVEEALAHPYL 410
GFLNENAKRYIRQLP YRRQSFQEKFP VHPEAIDLVEKMLTFDPR+RITVE ALAHPYL
Sbjct: 288 GFLNENAKRYIRQLPLYRRQSFQEKFPHVHPEAIDLVEKMLTFDPRQRITVENALAHPYL 347
Query: 409 TSLHDISDEPVCMTPFSFDFEQHALSEEQMKELIYREALAFKP 281
TSLHDISDEPVC TPFSFDFEQHAL+EEQMKELIYREALAF P
Sbjct: 348 TSLHDISDEPVCTTPFSFDFEQHALTEEQMKELIYREALAFNP 390
>gb|AAF65766.1|AF242308_1 mitogen-activated protein kinase [Euphorbia esula]
Length = 389
Score = 191 bits (486), Expect = 5e-48
Identities = 92/103 (89%), Positives = 97/103 (93%)
Frame = -1
Query: 589 GFLNENAKRYIRQLPPYRRQSFQEKFPQVHPEAIDLVEKMLTFDPRKRITVEEALAHPYL 410
GFLNENAKRYIRQ+ +RRQSF EKFP VHP AIDLVEKMLTFDPR+RITVE+ALAHPYL
Sbjct: 282 GFLNENAKRYIRQMSVFRRQSFTEKFPTVHPAAIDLVEKMLTFDPRQRITVEDALAHPYL 341
Query: 409 TSLHDISDEPVCMTPFSFDFEQHALSEEQMKELIYREALAFKP 281
TSLHDISDEPVCMTPFSFDFEQHAL+EEQMKELIYREALAF P
Sbjct: 342 TSLHDISDEPVCMTPFSFDFEQHALTEEQMKELIYREALAFNP 384
>gb|AAB58396.1| salicylic acid-activated MAP kinase [Nicotiana tabacum]
Length = 393
Score = 190 bits (482), Expect = 1e-47
Identities = 91/102 (89%), Positives = 95/102 (92%)
Frame = -1
Query: 586 FLNENAKRYIRQLPPYRRQSFQEKFPQVHPEAIDLVEKMLTFDPRKRITVEEALAHPYLT 407
FLNENAKRYIRQLP YRRQSF EKFP VHP AIDLVEKMLTFDPR+RITVE ALAHPYL
Sbjct: 287 FLNENAKRYIRQLPLYRRQSFTEKFPHVHPTAIDLVEKMLTFDPRRRITVEGALAHPYLN 346
Query: 406 SLHDISDEPVCMTPFSFDFEQHALSEEQMKELIYREALAFKP 281
SLHDISDEP+CMTPFSFDFEQHAL+EEQMKELIYRE+LAF P
Sbjct: 347 SLHDISDEPICMTPFSFDFEQHALTEEQMKELIYRESLAFNP 388
>sp|Q40532|NTF4_TOBAC MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG NTF4 (P45)
gi|1076640|pir||S51321 mitogen-activated protein kinase
4 (EC 2.7.1.-) - common tobacco
gi|634070|emb|CAA58761.1| p45Ntf4 serine/threonine
protein kinase [Nicotiana tabacum]
Length = 393
Score = 189 bits (480), Expect = 2e-47
Identities = 91/102 (89%), Positives = 96/102 (93%)
Frame = -1
Query: 586 FLNENAKRYIRQLPPYRRQSFQEKFPQVHPEAIDLVEKMLTFDPRKRITVEEALAHPYLT 407
FLNENAKRYIRQLP YRRQSF EKFP V+P AIDLVEKMLTFDPR+RITVE+ALAHPYLT
Sbjct: 287 FLNENAKRYIRQLPLYRRQSFVEKFPHVNPAAIDLVEKMLTFDPRRRITVEDALAHPYLT 346
Query: 406 SLHDISDEPVCMTPFSFDFEQHALSEEQMKELIYREALAFKP 281
SLHDISDEPVCMTPF+FDFEQHAL+EEQMKELIYRE LAF P
Sbjct: 347 SLHDISDEPVCMTPFNFDFEQHALTEEQMKELIYREGLAFNP 388
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 496,903,129
Number of Sequences: 1393205
Number of extensions: 10404464
Number of successful extensions: 29751
Number of sequences better than 10.0: 1814
Number of HSP's better than 10.0 without gapping: 28570
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29584
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)