Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
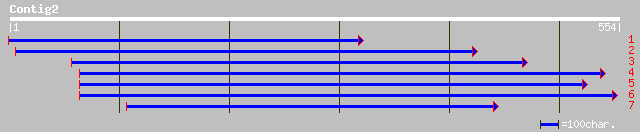
Query= KMC002970A_C02 KMC002970A_c02
(545 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_192837.1| putative retrotransposon polyprotein; protein i... 91 7e-18
pir||T01879 hypothetical protein F8M12.17 - Arabidopsis thaliana... 89 3e-17
dbj|BAB11447.1| polyprotein-like [Arabidopsis thaliana] 85 5e-16
ref|NP_193182.1| retrovirus-related like polyprotein; protein id... 84 1e-15
pir||T14518 hypothetical protein 2 - wild cabbage transposon Mel... 83 2e-15
>ref|NP_192837.1| putative retrotransposon polyprotein; protein id: At4g10990.1
[Arabidopsis thaliana] gi|7486142|pir||T04294
hypothetical protein F25I24.200 - Arabidopsis thaliana
gi|4539373|emb|CAB40067.1| putative retrotransposon
polyprotein [Arabidopsis thaliana]
gi|7267797|emb|CAB81200.1| putative retrotransposon
polyprotein [Arabidopsis thaliana]
Length = 1203
Score = 91.3 bits (225), Expect = 7e-18
Identities = 54/142 (38%), Positives = 79/142 (55%), Gaps = 14/142 (9%)
Frame = -3
Query: 528 CEALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFG 349
CE +WL L DL + ++CDN+SALHLA N VFH+RT++IEIDCH V ++ G
Sbjct: 938 CEIIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEIDCHTVRDQIKAG 997
Query: 348 ILHLLHVPSSDQVADVFTKTIS-----------QFTG---AISIPGCQSWIA*SPFLFGI 211
L LHVP+ +Q+AD+ TK + +FTG + + ++W+ S + +
Sbjct: 998 KLKTLHVPTGNQLADILTKPLHPVQSPIFSLLFRFTGTSPVLIVEAAEAWMP-SVAMSIV 1056
Query: 210 TDISLVSYFVILLLSVSPFLLN 145
DIS VS V ++S LN
Sbjct: 1057 VDISRVSDSVAQNANLSRSWLN 1078
>pir||T01879 hypothetical protein F8M12.17 - Arabidopsis thaliana
gi|3513747|gb|AAC33963.1| contains similarity to reverse
transcriptases (Pfam; rvt.hmm, score: 11.19) [Arabidopsis
thaliana]
Length = 1633
Score = 89.0 bits (219), Expect = 3e-17
Identities = 40/81 (49%), Positives = 55/81 (67%)
Frame = -3
Query: 528 CEALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFG 349
CE +WL L DL + ++CDN+SALHLA N VFH+RT++IEIDCH V ++ G
Sbjct: 1336 CEIIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEIDCHTVRDQIKAG 1395
Query: 348 ILHLLHVPSSDQVADVFTKTI 286
L LHVP+ +Q+AD+ TK +
Sbjct: 1396 KLKTLHVPTGNQLADILTKPL 1416
>dbj|BAB11447.1| polyprotein-like [Arabidopsis thaliana]
Length = 509
Score = 85.1 bits (209), Expect = 5e-16
Identities = 38/80 (47%), Positives = 55/80 (68%)
Frame = -3
Query: 525 EALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFGI 346
E LWL L DL + V ++CDN+SA+H+A NSVFH+RT+++EIDCH +V G
Sbjct: 400 ELLWLAQMLKDLHVEMEFQVKLFCDNKSAMHIANNSVFHERTKHVEIDCHTTRDRVKNGF 459
Query: 345 LHLLHVPSSDQVADVFTKTI 286
L +LHV + +Q+AD+ TK +
Sbjct: 460 LKVLHVDTENQLADILTKAL 479
>ref|NP_193182.1| retrovirus-related like polyprotein; protein id: At4g14460.1
[Arabidopsis thaliana] gi|7488175|pir||G71406 probable
retrovirus-related polyprotein - Arabidopsis thaliana
gi|2244802|emb|CAB10225.1| retrovirus-related like
polyprotein [Arabidopsis thaliana]
gi|7268152|emb|CAB78488.1| retrovirus-related like
polyprotein [Arabidopsis thaliana]
Length = 1489
Score = 83.6 bits (205), Expect = 1e-15
Identities = 39/81 (48%), Positives = 53/81 (65%)
Frame = -3
Query: 528 CEALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFG 349
CE +WL L DL I ++CDN+SALH + N VFH+RT++IEIDCH V ++ G
Sbjct: 1383 CEIIWLQQLLKDLHIPLTCPAKLFCDNKSALHSSLNPVFHERTKHIEIDCHTVRDQIKAG 1442
Query: 348 ILHLLHVPSSDQVADVFTKTI 286
L LHVP+ +Q AD+ TK +
Sbjct: 1443 NLKALHVPTENQHADILTKAL 1463
>pir||T14518 hypothetical protein 2 - wild cabbage transposon Melmoth
gi|2462936|emb|CAA72990.1| open reading frame 2
[Brassica oleracea]
Length = 253
Score = 82.8 bits (203), Expect = 2e-15
Identities = 37/81 (45%), Positives = 57/81 (69%)
Frame = -3
Query: 528 CEALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFG 349
CE +WL L +L +AS + V++ D+ +A+++A N VFH+RT++IE+DCH V K+ G
Sbjct: 152 CEMMWLCILLRELHVASSSVPVLFSDSTAAIYIATNPVFHERTKHIELDCHTVREKIDKG 211
Query: 348 ILHLLHVPSSDQVADVFTKTI 286
+L LHV + DQVAD+ TK +
Sbjct: 212 LLKTLHVRTEDQVADILTKPL 232
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 416,509,442
Number of Sequences: 1393205
Number of extensions: 8116893
Number of successful extensions: 23394
Number of sequences better than 10.0: 323
Number of HSP's better than 10.0 without gapping: 23042
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23381
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18660035355
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)