Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002969A_C01 KMC002969A_c01
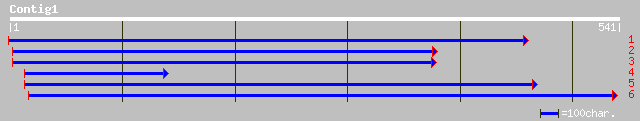
(540 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM10951.1|AF488595_1 putative bHLH transcription factor [Ara... 38 0.069
ref|NP_187390.1| bHLH protein; protein id: At3g07340.1, supporte... 38 0.069
ref|NP_496637.1| Predicted CDS, serpentine receptor Sre, putativ... 34 1.3
ref|NP_251731.1| hypothetical protein [Pseudomonas aeruginosa PA... 32 4.9
ref|NP_508914.1| Predicted CDS, putative cytoplasmic protein, wi... 32 6.4
>gb|AAM10951.1|AF488595_1 putative bHLH transcription factor [Arabidopsis thaliana]
Length = 450
Score = 38.1 bits (87), Expect = 0.069
Identities = 21/39 (53%), Positives = 24/39 (60%), Gaps = 2/39 (5%)
Frame = -3
Query: 490 TFSEDDLHTIVQMGFGQTANRKTPIRFQSIN--GSNQVP 380
TFSEDDLH+I+ MGF Q R Q +N SNQVP
Sbjct: 412 TFSEDDLHSIIHMGFAQN-------RLQELNQGSSNQVP 443
>ref|NP_187390.1| bHLH protein; protein id: At3g07340.1, supported by cDNA:
gi_20127063 [Arabidopsis thaliana]
gi|6041855|gb|AAF02164.1|AC009853_24 unknown protein
[Arabidopsis thaliana]
Length = 456
Score = 38.1 bits (87), Expect = 0.069
Identities = 21/39 (53%), Positives = 24/39 (60%), Gaps = 2/39 (5%)
Frame = -3
Query: 490 TFSEDDLHTIVQMGFGQTANRKTPIRFQSIN--GSNQVP 380
TFSEDDLH+I+ MGF Q R Q +N SNQVP
Sbjct: 418 TFSEDDLHSIIHMGFAQN-------RLQELNQGSSNQVP 449
>ref|NP_496637.1| Predicted CDS, serpentine receptor Sre, putative G protein-coupled
chemoreceptor family member [Caenorhabditis elegans]
gi|7510263|pir||T27200 hypothetical protein Y57A10C.4 -
Caenorhabditis elegans gi|3881140|emb|CAA19546.1|
Hypothetical protein Y57A10C.4 [Caenorhabditis elegans]
Length = 359
Score = 33.9 bits (76), Expect = 1.3
Identities = 20/48 (41%), Positives = 31/48 (63%), Gaps = 5/48 (10%)
Frame = -2
Query: 206 LAIPISFLFL---LTLSLSMIIIFNHLGLFF--SIFFTVKSLGTILYY 78
L IPISF+ + L +S+S+ I+FN LGL+ + + +GTI+YY
Sbjct: 166 LLIPISFIIIYQFLAISISLGILFNILGLYVLNASWILCILIGTIMYY 213
>ref|NP_251731.1| hypothetical protein [Pseudomonas aeruginosa PA01]
gi|11349769|pir||B83265 hypothetical protein PA3041
[imported] - Pseudomonas aeruginosa (strain PAO1)
gi|9949146|gb|AAG06429.1|AE004729_3 hypothetical protein
[Pseudomonas aeruginosa PAO1]
Length = 126
Score = 32.0 bits (71), Expect = 4.9
Identities = 27/75 (36%), Positives = 39/75 (52%), Gaps = 7/75 (9%)
Frame = -2
Query: 206 LAIPISFLFLLTLSLSMIIIF---NHL--GLFFSIFFTVKSL--GTILYYSLGANSCYYC 48
LA+ + L L+ LSL ++I+F N L L +F+ + SL G LY S+ S +
Sbjct: 52 LALVFALLLLVGLSLLVLIVFWDTNRLAAALGLCLFYVIGSLFCGWRLYQSINDESSPFS 111
Query: 47 AKL*DSAN*RSFFLP 3
A L + AN R LP
Sbjct: 112 ATLEELANDRERLLP 126
>ref|NP_508914.1| Predicted CDS, putative cytoplasmic protein, with a coiled coil-4
domain, nematode specific [Caenorhabditis elegans]
gi|7500488|pir||T16257 hypothetical protein F35C8.8 -
Caenorhabditis elegans gi|1072190|gb|AAA81712.1|
Hypothetical protein F35C8.8 [Caenorhabditis elegans]
Length = 312
Score = 31.6 bits (70), Expect = 6.4
Identities = 25/60 (41%), Positives = 30/60 (49%), Gaps = 5/60 (8%)
Frame = -2
Query: 236 GLIEASFVFMLAIPISFLFLLTLSLSM----IIIFNHLG-LFFSIFFTVKSLGTILYYSL 72
G IE VFM + L L TLSLS II G L F FF +KS T++Y +L
Sbjct: 137 GKIEGVIVFMYFSSSTLLLLHTLSLSFSWCHFIISGSCGKLTFDFFFLLKSNYTVVYRTL 196
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 444,758,732
Number of Sequences: 1393205
Number of extensions: 9372721
Number of successful extensions: 27170
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 24561
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26938
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)