Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
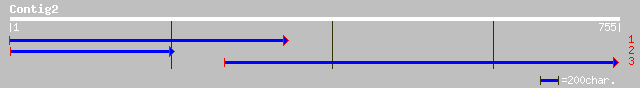
Query= KMC002968A_C02 KMC002968A_c02
(755 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM91610.1| putative protein [Arabidopsis thaliana] gi|231977... 43 2e-05
ref|NP_196259.1| putative protein; protein id: At5g06410.1 [Arab... 43 2e-05
emb|CAD53214.1| hypothetical protein, unlikely [Trypanosoma brucei] 35 1.5
ref|NP_179898.1| unknown protein; protein id: At2g23170.1, suppo... 33 3.4
ref|NP_288124.1| putative transport system permease protein [Esc... 33 5.8
>gb|AAM91610.1| putative protein [Arabidopsis thaliana] gi|23197714|gb|AAN15384.1|
putative protein [Arabidopsis thaliana]
Length = 252
Score = 43.1 bits (100), Expect(2) = 2e-05
Identities = 20/29 (68%), Positives = 27/29 (92%)
Frame = -2
Query: 751 EIMEIREAVAEANNSEALNQILSQMQEKL 665
EIME+REA++EA++S +LNQI SQ+QEKL
Sbjct: 184 EIMELREAISEADDSTSLNQIRSQVQEKL 212
Score = 27.3 bits (59), Expect(2) = 2e-05
Identities = 10/17 (58%), Positives = 14/17 (81%)
Frame = -1
Query: 593 RRMTYYSRVIDEVVKKL 543
+RMTYY R +E++KKL
Sbjct: 236 QRMTYYERACEEILKKL 252
>ref|NP_196259.1| putative protein; protein id: At5g06410.1 [Arabidopsis thaliana]
gi|9758421|dbj|BAB08963.1| gene_id:MHF15.7~similar to
unknown protein~sp|P36540 [Arabidopsis thaliana]
Length = 246
Score = 43.1 bits (100), Expect(2) = 2e-05
Identities = 20/29 (68%), Positives = 27/29 (92%)
Frame = -2
Query: 751 EIMEIREAVAEANNSEALNQILSQMQEKL 665
EIME+REA++EA++S +LNQI SQ+QEKL
Sbjct: 178 EIMELREAISEADDSTSLNQIRSQVQEKL 206
Score = 27.3 bits (59), Expect(2) = 2e-05
Identities = 10/17 (58%), Positives = 14/17 (81%)
Frame = -1
Query: 593 RRMTYYSRVIDEVVKKL 543
+RMTYY R +E++KKL
Sbjct: 230 QRMTYYERACEEILKKL 246
>emb|CAD53214.1| hypothetical protein, unlikely [Trypanosoma brucei]
Length = 167
Score = 34.7 bits (78), Expect = 1.5
Identities = 26/94 (27%), Positives = 42/94 (44%)
Frame = -3
Query: 492 GFHSQVYSSNTYTSTDMFFLYNLCNVVHSFILRCPYL*FCL*FLYCHFIIDYISVGKDGV 313
G H +V +N+Y + L L + H +L P CL YC + I D
Sbjct: 29 GSHKRVRENNSYIIPLLNVLRPLFS--HLLVLFPPSFSLCLPPPYCSPMFYKIPASMDTR 86
Query: 312 IPLD*SHLRMKWPLRRVPYDLIFLSTPFCFSMHI 211
+ L + R+ +P + +P+ + LSTP F H+
Sbjct: 87 LVLVFTRFRVVFPRKLIPFFVHLLSTPSLFHYHV 120
>ref|NP_179898.1| unknown protein; protein id: At2g23170.1, supported by cDNA:
gi_19699327 [Arabidopsis thaliana]
gi|7487246|pir||T00515 auxin-regulated protein GH3
homolog At2g23170 - Arabidopsis thaliana
gi|2642446|gb|AAB87114.1| unknown protein [Arabidopsis
thaliana] gi|19699328|gb|AAL91274.1| At2g23170/T20D16.20
[Arabidopsis thaliana] gi|23463057|gb|AAN33198.1|
At2g23170/T20D16.20 [Arabidopsis thaliana]
Length = 595
Score = 33.5 bits (75), Expect = 3.4
Identities = 22/66 (33%), Positives = 33/66 (49%)
Frame = -2
Query: 376 LPLIPLLSFYY*LYQCRQRRSDSFGLITSPNEVASQKSTI*SHISQYTFLLLYAHFVTS* 197
LP P+L+ YY Q ++R +D + + TSPNE + S +Q LL H V
Sbjct: 167 LPARPVLTSYYKSEQFKRRPNDPYNVYTSPNEAILCPDSSQSMYTQMLCGLLMRHEVLRL 226
Query: 196 GRLHAT 179
G + A+
Sbjct: 227 GAVFAS 232
>ref|NP_288124.1| putative transport system permease protein [Escherichia coli
O157:H7 EDL933] gi|15831651|ref|NP_310424.1| putative
transport system permease protein [Escherichia coli
O157:H7] gi|25303025|pir||E90928 probable transport
system permease protein ECs2397 [imported] - Escherichia
coli (strain O157:H7, substrain RIMD 0509952)
gi|25303027|pir||A85777 probable transport system
permease protein Z2718 [imported] - Escherichia coli
(strain O157:H7, substrain EDL933)
gi|12515695|gb|AAG56677.1|AE005392_3 putative transport
system permease protein [Escherichia coli O157:H7
EDL933] gi|13361864|dbj|BAB35820.1| putative transport
system permease protein [Escherichia coli O157:H7]
Length = 404
Score = 32.7 bits (73), Expect = 5.8
Identities = 15/38 (39%), Positives = 22/38 (57%)
Frame = -1
Query: 536 RTSLLLTLFCLLHFKAFTVKCIHPTLILLLICSFSITF 423
R + LL L+ + F A C+HPT +++I SF I F
Sbjct: 273 RPTTLLMLYTFISFIALFTVCLHPTFYVVIIFSFVIGF 310
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 578,553,104
Number of Sequences: 1393205
Number of extensions: 11489038
Number of successful extensions: 24160
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 23497
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24151
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 36877108757
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)