Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
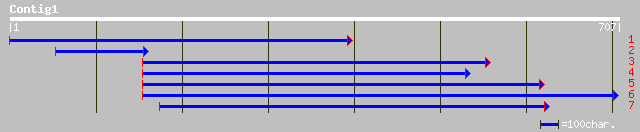
Query= KMC002960A_C01 KMC002960A_c01
(705 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_566370.1| expressed protein; protein id: At3g10250.1, sup... 119 3e-26
gb|AAF02817.1|AC009400_13 unknown protein [Arabidopsis thaliana] 119 3e-26
ref|NP_196029.1| putative protein; protein id: At5g04090.1 [Arab... 100 2e-20
pir||T48436 hypothetical protein F8F6.300 - Arabidopsis thaliana... 100 2e-20
pir||F84902 hypothetical protein At2g46420 [imported] - Arabidop... 36 0.45
>ref|NP_566370.1| expressed protein; protein id: At3g10250.1, supported by cDNA:
106330., supported by cDNA: gi_15809981, supported by
cDNA: gi_18958053 [Arabidopsis thaliana]
gi|15809982|gb|AAL06918.1| AT3g10250/F14P13_15
[Arabidopsis thaliana] gi|18958054|gb|AAL79600.1|
AT3g10250/F14P13_15 [Arabidopsis thaliana]
gi|21536630|gb|AAM60962.1| unknown [Arabidopsis
thaliana]
Length = 324
Score = 119 bits (299), Expect = 3e-26
Identities = 60/126 (47%), Positives = 92/126 (72%), Gaps = 1/126 (0%)
Frame = -1
Query: 705 MRSAQSSSMGLIQGINGGMIKSEPGYSGSPSYIFGPDSTVLDACPTIGDGTVTSFTSVNS 526
M S+Q+++M ++QG+NGGMIKSE ++ S+++G + L+ +GD ++ +F++ S
Sbjct: 176 MLSSQTTNMPMMQGMNGGMIKSETAFTNPASFMYGGERNALEGHSAVGDTSIPNFSN-ES 234
Query: 525 NSHSMNGALLDLDISSFGVLGQISRNLSLSDLTADFSQSSDIMESYPRCPYLATN-ENFL 349
N+ ++ LL+ + S+FG LGQI RN SLSDLTADFSQSS+I+ESY R P+L N ENFL
Sbjct: 235 NNQPLSDPLLEAEASTFGFLGQIPRNFSLSDLTADFSQSSEILESYDRSPFLVPNAENFL 294
Query: 348 ES*EQG 331
+S ++G
Sbjct: 295 DSRDRG 300
>gb|AAF02817.1|AC009400_13 unknown protein [Arabidopsis thaliana]
Length = 327
Score = 119 bits (299), Expect = 3e-26
Identities = 60/126 (47%), Positives = 92/126 (72%), Gaps = 1/126 (0%)
Frame = -1
Query: 705 MRSAQSSSMGLIQGINGGMIKSEPGYSGSPSYIFGPDSTVLDACPTIGDGTVTSFTSVNS 526
M S+Q+++M ++QG+NGGMIKSE ++ S+++G + L+ +GD ++ +F++ S
Sbjct: 179 MLSSQTTNMPMMQGMNGGMIKSETAFTNPASFMYGGERNALEGHSAVGDTSIPNFSN-ES 237
Query: 525 NSHSMNGALLDLDISSFGVLGQISRNLSLSDLTADFSQSSDIMESYPRCPYLATN-ENFL 349
N+ ++ LL+ + S+FG LGQI RN SLSDLTADFSQSS+I+ESY R P+L N ENFL
Sbjct: 238 NNQPLSDPLLEAEASTFGFLGQIPRNFSLSDLTADFSQSSEILESYDRSPFLVPNAENFL 297
Query: 348 ES*EQG 331
+S ++G
Sbjct: 298 DSRDRG 303
>ref|NP_196029.1| putative protein; protein id: At5g04090.1 [Arabidopsis thaliana]
gi|9955564|emb|CAC05491.1| putative protein [Arabidopsis
thaliana]
Length = 300
Score = 100 bits (249), Expect = 2e-20
Identities = 62/123 (50%), Positives = 82/123 (66%), Gaps = 1/123 (0%)
Frame = -1
Query: 699 SAQSSSMGLIQGINGGMIKSEPGYSGSPSYIFGPDSTVLDACPTIGDGTVTSFTSVNSNS 520
S Q+++M L+QG MIKSE Y Y++G + A T+GD T+ SF++ +SN
Sbjct: 177 SLQATNMPLMQG----MIKSETAYQNCAPYMYGGE-----AQSTVGDVTIASFSNDSSNQ 227
Query: 519 HSMNGALLDLDISSFGVLGQISRNLSLSDLTADFSQSSDIMESYPRCPY-LATNENFLES 343
S+N L+D D +FG LGQI +N SLSDLTADFSQSSDI+ESY P+ LA ENFL+S
Sbjct: 228 -SLNDPLVDPDAPTFGSLGQIPQNFSLSDLTADFSQSSDILESYEGSPFLLADAENFLDS 286
Query: 342 *EQ 334
E+
Sbjct: 287 SER 289
>pir||T48436 hypothetical protein F8F6.300 - Arabidopsis thaliana (fragment)
gi|7406419|emb|CAB85529.1| putative protein [Arabidopsis
thaliana]
Length = 275
Score = 100 bits (249), Expect = 2e-20
Identities = 62/123 (50%), Positives = 82/123 (66%), Gaps = 1/123 (0%)
Frame = -1
Query: 699 SAQSSSMGLIQGINGGMIKSEPGYSGSPSYIFGPDSTVLDACPTIGDGTVTSFTSVNSNS 520
S Q+++M L+QG MIKSE Y Y++G + A T+GD T+ SF++ +SN
Sbjct: 152 SLQATNMPLMQG----MIKSETAYQNCAPYMYGGE-----AQSTVGDVTIASFSNDSSNQ 202
Query: 519 HSMNGALLDLDISSFGVLGQISRNLSLSDLTADFSQSSDIMESYPRCPY-LATNENFLES 343
S+N L+D D +FG LGQI +N SLSDLTADFSQSSDI+ESY P+ LA ENFL+S
Sbjct: 203 -SLNDPLVDPDAPTFGSLGQIPQNFSLSDLTADFSQSSDILESYEGSPFLLADAENFLDS 261
Query: 342 *EQ 334
E+
Sbjct: 262 SER 264
>pir||F84902 hypothetical protein At2g46420 [imported] - Arabidopsis thaliana
Length = 279
Score = 36.2 bits (82), Expect = 0.45
Identities = 35/93 (37%), Positives = 45/93 (47%), Gaps = 3/93 (3%)
Frame = -1
Query: 603 GPDSTVLDACPTIGDGTVTSFTSVNSNSHSMNGALLDLDISSFGVLGQISRNLSLSDLTA 424
G D++ LD+ T GT SV S GA D QI N SLSDLTA
Sbjct: 188 GMDTSALDSAFTSDVGT-----SVGLQLGSDGGAGNSRD--PLRPFDQIPWNFSLSDLTA 240
Query: 423 DFSQSSDI--MESYPRCPYL-ATNENFLES*EQ 334
D S D+ + +YP P+L + +E L+S EQ
Sbjct: 241 DLSNLGDLGALGNYPGSPFLPSDSEILLDSPEQ 273
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 551,079,638
Number of Sequences: 1393205
Number of extensions: 11341768
Number of successful extensions: 28306
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 26966
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28271
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32091529758
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)